2KC8
 
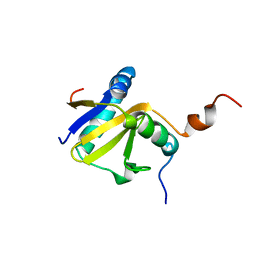 | | Structure of E. coli toxin RelE (R81A/R83A) mutant in complex with antitoxin RelBc (K47-L79) peptide | | Descriptor: | Antitoxin RelB, Toxin relE | | Authors: | Li, G, Zhang, Y, Inouye, M, Ikura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-03-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site.
J.Biol.Chem., 284, 2009
|
|
5UAE
 
 | | Crystal structure of the coiled-coil domain from Listeria Innocua Phage Integrase (Trigonal Form) | | Descriptor: | CITRATE ANION, Putative integrase | | Authors: | Gupta, K, Sharp, R, Yuan, J.B, Van Duyne, G.D. | | Deposit date: | 2016-12-19 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Coiled-coil interactions mediate serine integrase directionality.
Nucleic Acids Res., 45, 2017
|
|
5UDO
 
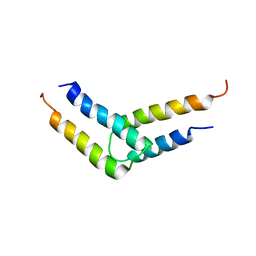 | |
2KAA
 
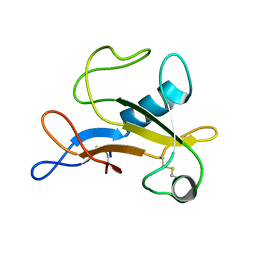 | |
2KC9
 
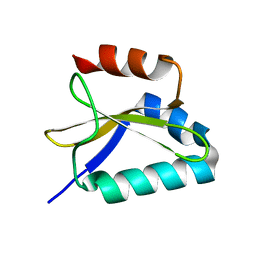 | |
8I8R
 
 | | Cryo-EM Structure of OmpC3-MlaA Complex in MSP2N2 Nanodiscs | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Intermembrane phospholipid transport system lipoprotein MlaA, Outer membrane porin C | | Authors: | Yeow, J, Luo, M, Chng, S.S. | | Deposit date: | 2023-02-05 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Molecular mechanism of phospholipid transport at the bacterial outer membrane interface.
Nat Commun, 14, 2023
|
|
8I8X
 
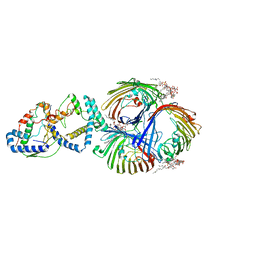 | | Cryo-EM Structure of OmpC3-MlaA-MlaC Complex in MSP2N2 Nanodiscs | | Descriptor: | (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-[(2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-2-carboxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-5-[[(3~{R})-3-dodecanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanyltetradecanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-4-[(3~{R})-3-tetradecanoyloxytetradecanoyl]oxy-oxan-2-yl]methoxy]-5-oxidanyl-oxan-4-yl]oxy-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Intermembrane phospholipid transport system binding protein MlaC, Intermembrane phospholipid transport system lipoprotein MlaA, ... | | Authors: | Yeow, J, Luo, M, Chng, S.S. | | Deposit date: | 2023-02-05 | | Release date: | 2023-12-20 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Molecular mechanism of phospholipid transport at the bacterial outer membrane interface.
Nat Commun, 14, 2023
|
|
6FLY
 
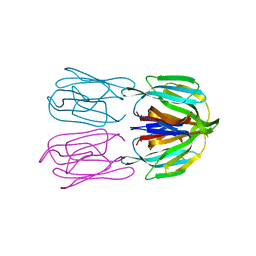 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with mannose. | | Descriptor: | Jacalin-like lectin, alpha-D-mannopyranose | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.749 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
6G5P
 
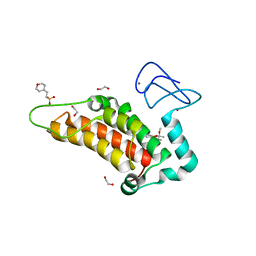 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment FM009493b 2,3-Dimethoxy-2,3-dimethyl-2,3-dihydro-1,4-benzodioxin-6-amine | | Descriptor: | (2~{R},3~{R})-2,3-dimethoxy-2,3-dimethyl-1,4-benzodioxin-6-amine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
6FLZ
 
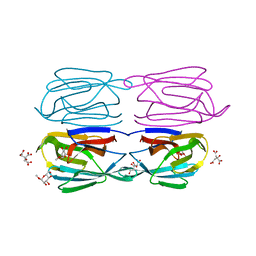 | | Structure of AcmJRL, a mannose binding jacalin related lectin from Ananas comosus, in complex with methyl-mannose. | | Descriptor: | CITRIC ACID, Jacalin-like lectin, methyl alpha-D-mannopyranoside | | Authors: | Azarkan, M, Herman, R, El Mahyaoui, R, Sauvage, E, Vanden Broeck, A, Charlier, P. | | Deposit date: | 2018-01-29 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Biochemical and structural characterization of a mannose binding jacalin-related lectin with two-sugar binding sites from pineapple (Ananas comosus) stem.
Sci Rep, 8, 2018
|
|
2KO2
 
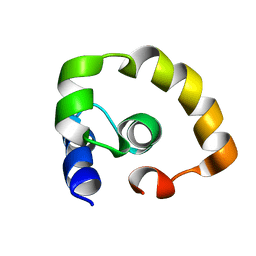 | | NOGO66 | | Descriptor: | Reticulon-4 | | Authors: | Cocco, M.J, Schulz, J, Vasudevan, S.V. | | Deposit date: | 2009-09-08 | | Release date: | 2010-04-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Protein folding at the membrane interface, the structure of Nogo-66 requires interactions with a phosphocholine surface.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7PD3
 
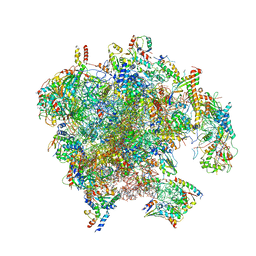 | | Structure of the human mitoribosomal large subunit in complex with NSUN4.MTERF4.GTPBP7 and MALSU1.L0R8F8.mt-ACP | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Chandrasekaran, V, Desai, N, Burton, N.O, Yang, H, Price, J, Miska, E.A, Ramakrishnan, V. | | Deposit date: | 2021-08-04 | | Release date: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Visualizing formation of the active site in the mitochondrial ribosome.
Elife, 10, 2021
|
|
1NZ6
 
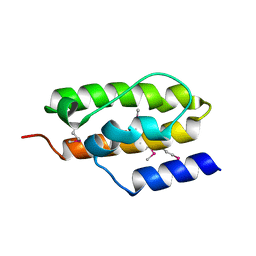 | | Crystal Structure of Auxilin J-Domain | | Descriptor: | Auxilin | | Authors: | Jiang, J, Taylor, A.B, Prasad, K, Ishikawa-Brush, Y, Hart, P.J, Lafer, E.M, Sousa, R. | | Deposit date: | 2003-02-16 | | Release date: | 2003-04-22 | | Last modified: | 2011-11-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface.
Biochemistry, 42, 2003
|
|
3QBQ
 
 | | Crystal structure of extracellular domains of mouse RANK-RANKL complex | | Descriptor: | Tumor necrosis factor ligand superfamily member 11, Tumor necrosis factor receptor superfamily member 11A | | Authors: | Ta, H.M, Nguyen, G.T.T, Jin, H.M, Choi, J.K, Park, H, Kim, N.S, Hwang, H.Y, Kim, K.K. | | Deposit date: | 2011-01-13 | | Release date: | 2011-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based development of a receptor activator of nuclear factor-kappaB ligand (RANKL) inhibitor peptide and molecular basis for osteopetrosis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1D6R
 
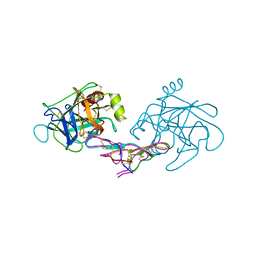 | | CRYSTAL STRUCTURE OF CANCER CHEMOPREVENTIVE BOWMAN-BIRK INHIBITOR IN TERNARY COMPLEX WITH BOVINE TRYPSIN AT 2.3 A RESOLUTION. STRUCTURAL BASIS OF JANUS-FACED SERINE PROTEASE INHIBITOR SPECIFICITY | | Descriptor: | BOWMAN-BIRK PROTEINASE INHIBITOR PRECURSOR, TRYPSINOGEN | | Authors: | Koepke, J, Ermler, U, Wenzl, G, Flecker, P. | | Deposit date: | 1999-10-15 | | Release date: | 2000-05-05 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cancer chemopreventive Bowman-Birk inhibitor in ternary complex with bovine trypsin at 2.3 A resolution. Structural basis of Janus-faced serine protease inhibitor specificity.
J.Mol.Biol., 298, 2000
|
|
1DN2
 
 | | FC FRAGMENT OF HUMAN IGG1 IN COMPLEX WITH AN ENGINEERED 13 RESIDUE PEPTIDE DCAWHLGELVWCT-NH2 | | Descriptor: | ENGINEERED PEPTIDE, IMMUNOGLOBULIN LAMBDA HEAVY CHAIN, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLano, W.L, Ultsch, M.H, de Vos, A.M, Wells, J.A. | | Deposit date: | 1999-12-15 | | Release date: | 2000-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Convergent solutions to binding at a protein-protein interface.
Science, 287, 2000
|
|
6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
2JK2
 
 | | STRUCTURAL BASIS OF HUMAN TRIOSEPHOSPHATE ISOMERASE DEFICIENCY. CRYSTAL STRUCTURE OF THE WILD TYPE ENZYME. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, De Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-06-22 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
8C7I
 
 | |
1CZ8
 
 | | VASCULAR ENDOTHELIAL GROWTH FACTOR IN COMPLEX WITH AN AFFINITY MATURED ANTIBODY | | Descriptor: | HEAVY CHAIN OF NEUTRALIZING ANTIBODY, LIGHT CHAIN OF NEUTRALIZING ANTIBODY, SULFATE ION, ... | | Authors: | Chen, Y, Wiesmann, C, Fuh, G, Li, B, Christinger, H.W, McKay, P, de Vos, A.M, Lowman, H.B. | | Deposit date: | 1999-09-01 | | Release date: | 2000-03-20 | | Last modified: | 2017-04-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selection and analysis of an optimized anti-VEGF antibody: crystal structure of an affinity-matured Fab in complex with antigen.
J.Mol.Biol., 293, 1999
|
|
3HPG
 
 | |
8DPX
 
 | | Preligand association structure of DR5 | | Descriptor: | Tumor necrosis factor receptor superfamily member 10B | | Authors: | Du, G, Zhao, L, Chou, J.J. | | Deposit date: | 2022-07-17 | | Release date: | 2023-02-15 | | Method: | SOLUTION NMR | | Cite: | Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation.
Cell Res., 33, 2023
|
|
8F21
 
 | | Structure of a 30mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-06 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
8F0U
 
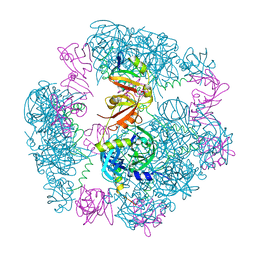 | | Structure of a 12mer DegP cage bound to the client protein hTRF1 | | Descriptor: | Periplasmic serine endoprotease DegP, Telomeric repeat-binding factor 1 | | Authors: | Harkness, R.W, Ripstein, Z.A, Di Trani, J.M, Kay, L.E. | | Deposit date: | 2022-11-04 | | Release date: | 2022-11-23 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Flexible Client-Dependent Cages in the Assembly Landscape of the Periplasmic Protease-Chaperone DegP.
J.Am.Chem.Soc., 145, 2023
|
|
