7M0A
 
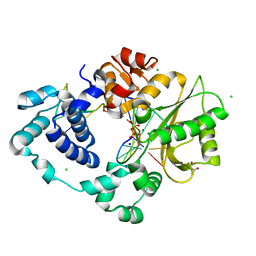 | | Incomplete in crystallo incorporation by DNA Polymerase Lambda bound to blunt-ended DSB substrate and incoming dTTP | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*AP*GP*TP*GP*CP*T)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
2JXH
 
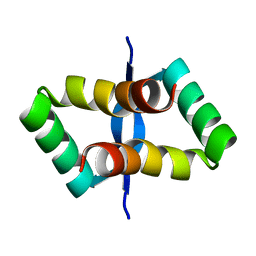 | |
3KA8
 
 | | Frog M-ferritin, EQH mutant, with cobalt | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferritin, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-19 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
3AOA
 
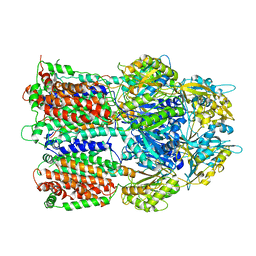 | |
2P1W
 
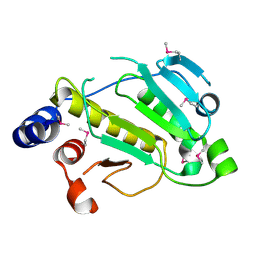 | |
7D4Q
 
 | | Structure of human TRPC5 in complex with HC-070 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 8-(3-chloranylphenoxy)-7-[(4-chlorophenyl)methyl]-3-methyl-1-(3-oxidanylpropyl)purine-2,6-dione, CALCIUM ION, ... | | Authors: | Chen, L, Song, K, Wei, M, Guo, W. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-31 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structural basis for human TRPC5 channel inhibition by two distinct inhibitors.
Elife, 10, 2021
|
|
7QZU
 
 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 47 | | Descriptor: | (2~{S})-2-[2-[4-[3,4-bis(oxidanyl)-9,10-bis(oxidanylidene)anthracen-2-yl]sulfonylpiperazin-1-yl]-2-oxidanylidene-ethyl]-2-oxidanyl-butanedioic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2022-01-31 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
7LZ5
 
 | |
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
3KA3
 
 | | Frog M-ferritin with magnesium | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Tosha, T, Ng, H.L, Theil, E, Alber, T, Bhattasali, O. | | Deposit date: | 2009-10-18 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites.
J.Am.Chem.Soc., 132, 2010
|
|
2PAH
 
 | |
7B9A
 
 | | CooS-V with Xe-soaked | | Descriptor: | 3,5-dioxa-7-thia-1-thionia-2$l^{2},4$l^{2},6$l^{3},8$l^{2}-tetraferrabicyclo[4.2.0]octane, BROMIDE ION, Carbon monoxide dehydrogenase, ... | | Authors: | Jeoung, J.H, Dobbek, H. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Morphing [4Fe-3S-nO]-Cluster within a Carbon Monoxide Dehydrogenase Scaffold.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
5M26
 
 | | Crystal structure of hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 in complex with methylhydroquinone | | Descriptor: | 2-methylbenzene-1,4-diol, FE (III) ION, Hydroquinone dioxygenase large subunit, ... | | Authors: | Ferraroni, M, Da Vela, S, Scozzafava, A, Kolvenbach, B, Corvini, P.F.X. | | Deposit date: | 2016-10-12 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of native hydroquinone 1,2-dioxygenase from Sphingomonas sp. TTNP3 and of substrate and inhibitor complexes.
Biochim. Biophys. Acta, 1865, 2017
|
|
7UAP
 
 | |
5M2W
 
 | | Structure of nanobody nb18 raised against TssK from E. coli T6SS | | Descriptor: | Llama nanobody nb8 against TssK from T6SS, SULFATE ION | | Authors: | Cambillau, C, Nguyen, V.S, Spinelli, S, Desmyter, A. | | Deposit date: | 2016-10-13 | | Release date: | 2017-06-28 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Type VI secretion TssK baseplate protein exhibits structural similarity with phage receptor-binding proteins and evolved to bind the membrane complex.
Nat Microbiol, 2, 2017
|
|
7BAQ
 
 | |
3J34
 
 | | Structure of HIV-1 Capsid Protein by Cryo-EM | | Descriptor: | capsid protein | | Authors: | Zhao, G, Perilla, J.R, Yufenyuy, E, Meng, X, Chen, B, Ning, J, Ahn, J, Gronenborn, A.M, Schulten, K, Aiken, C, Zhang, P. | | Deposit date: | 2013-02-23 | | Release date: | 2013-05-29 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (8.6 Å) | | Cite: | Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics.
Nature, 497, 2013
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
2OAG
 
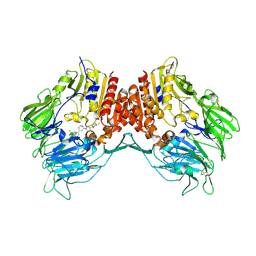 | | Crystal structure of human dipeptidyl peptidase IV (DPPIV) with pyrrolidine-constrained phenethylamine 29g | | Descriptor: | (3R,4S)-1-{6-[3-(METHYLSULFONYL)PHENYL]PYRIMIDIN-4-YL}-4-(2,4,5-TRIFLUOROPHENYL)PYRROLIDIN-3-AMINE, Dipeptidyl peptidase 4 | | Authors: | Backes, B.J, Longenecker, K.L, Hamilton, G.L, Stewart, K.D, Lai, C, Kopecka, H. | | Deposit date: | 2006-12-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pyrrolidine-constrained phenethylamines: The design of potent, selective, and pharmacologically efficacious dipeptidyl peptidase IV (DPP4) inhibitors from a lead-like screening hit.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5M38
 
 | |
7UAR
 
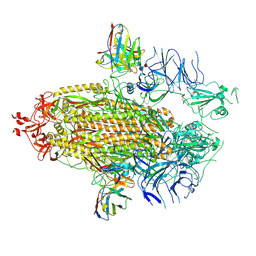 | |
7DL9
 
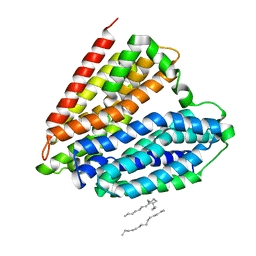 | | Crystal structure of nucleoside transporter NupG | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Nucleoside permease NupG | | Authors: | Wang, C, Xiao, Q.J, Deng, D. | | Deposit date: | 2020-11-26 | | Release date: | 2021-04-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for substrate recognition by the bacterial nucleoside transporter NupG.
J.Biol.Chem., 296, 2021
|
|
7UAQ
 
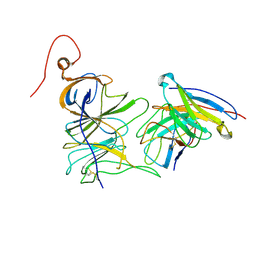 | | Structure of the SARS-CoV-2 NTD in complex with C1520, local refinement | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C1520 Fab Heavy Chain, ... | | Authors: | Barnes, C.O. | | Deposit date: | 2022-03-13 | | Release date: | 2022-04-27 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of memory B cells identifies conserved neutralizing epitopes on the N-terminal domain of variant SARS-Cov-2 spike proteins.
Immunity, 55, 2022
|
|
7DJ1
 
 | | Crystal structure of the G26C mutant of LeuT | | Descriptor: | LEUCINE, Na(+):neurotransmitter symporter (Snf family), SODIUM ION | | Authors: | Fan, J, Xiao, Y, Sun, Z, Zhou, X. | | Deposit date: | 2020-11-19 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.528 Å) | | Cite: | Crystal structures of LeuT reveal conformational dynamics in the outward-facing states.
J.Biol.Chem., 296, 2021
|
|
