7JVL
 
 | |
7FJ1
 
 | | Cryo-EM structure of pseudorabies virus C-capsid | | Descriptor: | Capsid vertex component 1, DNA packaging tegument protein UL25, Major capsid protein, ... | | Authors: | Zheng, Q, Li, S, Zha, Z, Sun, H. | | Deposit date: | 2021-08-02 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structures of pseudorabies virus capsids.
Nat Commun, 13, 2022
|
|
7QKE
 
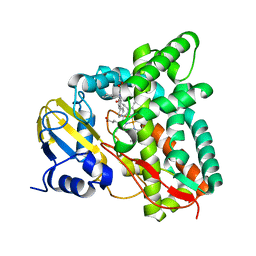 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclohexylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7C06
 
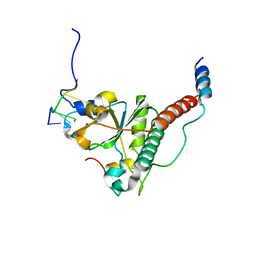 | | Crystal structure of yeast U2AF1 complex bound to 3' splice site RNA, 5'-UAGGU. | | Descriptor: | RNA (5'-R(*U*UP*AP*GP*GP*U)-3'), Splicing factor U2AF 23 kDa subunit, Splicing factor U2AF 59 kDa subunit, ... | | Authors: | Yoshida, H, Park, S.Y, Urano, T, Obayashi, E. | | Deposit date: | 2020-04-30 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Elucidation of the aberrant 3' splice site selection by cancer-associated mutations on the U2AF1.
Nat Commun, 11, 2020
|
|
2YO1
 
 | |
7JOT
 
 | |
7U5U
 
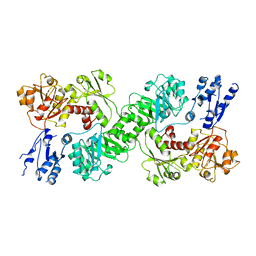 | |
2HZK
 
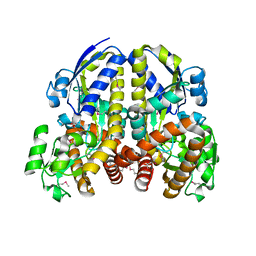 | | Crystal structures of a sodium-alpha-keto acid binding subunit from a TRAP transporter in its open form | | Descriptor: | GLYCEROL, TRAP-T family sorbitol/mannitol transporter, periplasmic binding protein, ... | | Authors: | Gonin, S, Arnoux, P, Pierru, B, Alonso, B, Sabaty, M, Pignol, D. | | Deposit date: | 2006-08-09 | | Release date: | 2007-04-03 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding.
Bmc Struct.Biol., 7, 2007
|
|
5M90
 
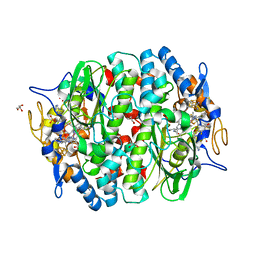 | | PCE reductive dehalogenase from S. multivorans in complex with 3,4,5-trifluorophenol | | Descriptor: | 3,4,5-tris(fluoranyl)phenol, BENZAMIDINE, GLYCEROL, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-31 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
2I05
 
 | | Escherichia Coli Replication Terminator Protein (Tus) Complexed With TerA DNA | | Descriptor: | 5'-D(*T*TP*AP*GP*TP*TP*AP*CP*AP*AP*CP*AP*TP*AP*CP*T)-3', 5'-D(*TP*AP*GP*TP*AP*TP*GP*TP*TP*GP*TP*AP*AP*CP*TP*A)-3', DNA replication terminus site-binding protein, ... | | Authors: | Oakley, A.J, Mulcair, M.D, Schaeffer, P.M, Dixon, N.E. | | Deposit date: | 2006-08-10 | | Release date: | 2007-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Polarity of Termination of DNA Replication in E. coli
To be Published
|
|
7QNN
 
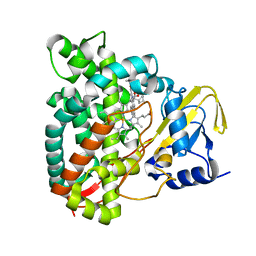 | | Crystal structure of CYP125 from Mycobacterium tuberculosis in complex with inhibitor (surface entropy reduction mutant) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Steroid C26-monooxygenase, ethyl 1-(cyclopentylmethyl)-5-pyridin-4-yl-indole-2-carboxylate | | Authors: | Snee, M, Tunnicliffe, R, Leys, D, Levy, C, Katariya, M. | | Deposit date: | 2021-12-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure Based Discovery of Inhibitors of CYP125 and CYP142 from Mycobacterium tuberculosis.
Chemistry, 29, 2023
|
|
7FFM
 
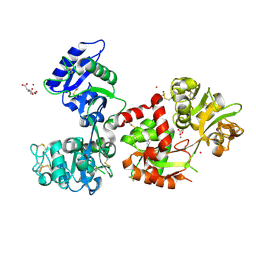 | | Human serum transferrin with five osmium binding sites | | Descriptor: | MALONATE ION, NITRILOTRIACETIC ACID, OSMIUM ION, ... | | Authors: | Wang, M, Sun, H. | | Deposit date: | 2021-07-23 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Binding of ruthenium and osmium at non‐iron sites of transferrin accounts for their iron-independent cellular uptake.
J.Inorg.Biochem., 234, 2022
|
|
2Y6T
 
 | | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia pestis | | Descriptor: | CHYMOTRYPSINOGEN A, ECOTIN, SULFATE ION | | Authors: | Clark, E.A, Walker, N, Ford, D.C, Cooper, I.A, Oyston, P.C.F, Acharya, K.R. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular Recognition of Chymotrypsin by the Serine Protease Inhibitor Ecotin from Yersinia Pestis.
J.Biol.Chem., 286, 2011
|
|
7U5S
 
 | |
3K70
 
 | |
7BSD
 
 | |
2YDE
 
 | | FACTOR INHIBITING HIF-1 ALPHA IN COMPLEX WITH S-2-HYDROXYGLUTARATE | | Descriptor: | (2S)-2-HYDROXYPENTANEDIOIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Chowdhury, R, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2011-03-18 | | Release date: | 2011-03-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases.
EMBO Rep., 12, 2011
|
|
7JQ1
 
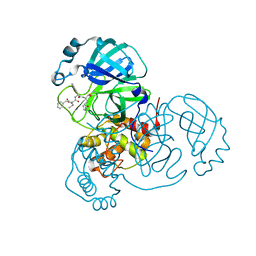 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI4 | | Descriptor: | 3C-like proteinase, N-[(benzyloxy)carbonyl]-L-valyl-N-{(2S)-1-hydroxy-3-[(3R)-2-oxo-3,4-dihydro-2H-pyrrol-3-yl]propan-2-yl}-L-phenylalaninamide | | Authors: | Yang, K, Liu, W. | | Deposit date: | 2020-08-10 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors*.
Chemmedchem, 16, 2021
|
|
7QSQ
 
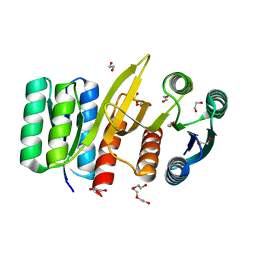 | | Permutated N-terminal lobe of the ribose binding protein from Thermotoga maritima | | Descriptor: | 1,2-ETHANEDIOL, Ribose ABC transporter, periplasmic ribose-binding protein, ... | | Authors: | Shanmugaratnam, S, Michel, F, Hocker, B. | | Deposit date: | 2022-01-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structures of permuted halves of a modern ribose-binding protein.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
2I1I
 
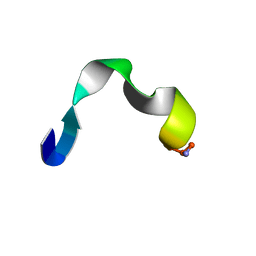 | | DPC micelle-bound NMR structures of Tritrp8 | | Descriptor: | 13-mer analogue of Prophenin-1 containing WWW | | Authors: | Schibli, D.J, Nguyen, L.T. | | Deposit date: | 2006-08-14 | | Release date: | 2006-11-28 | | Last modified: | 2020-06-24 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of tritrpticin analogs: potential relationships between antimicrobial activities, model membrane interactions, and their micelle-bound NMR structures
Biophys.J., 91, 2006
|
|
5MA0
 
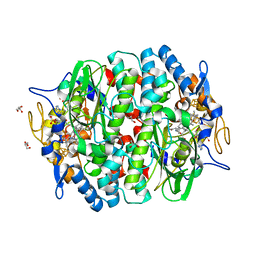 | | PCE reductive dehalogenase from S. multivorans in complex with 2,6-dichlorophenol | | Descriptor: | 2,6-dichlorophenol, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-11-02 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
5MMD
 
 | | TMB-1. Structural insights into TMB-1 and the role of residue 119 and 228 in substrate and inhibitor binding | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase 1, ZINC ION | | Authors: | Skagseth, S, Christopeit, T, Akhter, S, Bayer, A, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2016-12-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into TMB-1 and the Role of Residues 119 and 228 in Substrate and Inhibitor Binding.
Antimicrob. Agents Chemother., 61, 2017
|
|
7QNS
 
 | | Peptide VYEKKP in complex with human cathepsin V C25S mutant | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, Cathepsin L2, ... | | Authors: | Loboda, J, Sosnowski, P, Tusar, L, Vidmar, R, Vizovisek, M, Horvat, J, Kosec, G, Impens, F, Demol, H, Turk, B, Gevaert, K, Turk, D. | | Deposit date: | 2021-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide VYEKKP in complex with human cathepsin V C25S mutant
To Be Published
|
|
7FGX
 
 | | Toxoplasma gondii dihydrofolate reductase thymidylate synthase (TgDHFR-TS) complexed with P39, NADPH and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 6-hexyl-5-phenyl-pyrimidine-2,4-diamine, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Vanichtanankul, J, Yoomuang, A, Taweechai, S, Saeyang, T, Yuvaniyama, J, Tarnchompoo, B, Yuthavong, Y, Kamchonwongpaisan, S. | | Deposit date: | 2021-07-28 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into Effective Inhibitors' Binding to Toxoplasma gondii Dihydrofolate Reductase Thymidylate Synthase.
Acs Chem.Biol., 17, 2022
|
|
7BTC
 
 | | Crystal structure of Rheb Y35N mutant bound to GDP | | Descriptor: | GTP-binding protein Rheb, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Zhang, C, Zhang, T, Ding, J. | | Deposit date: | 2020-04-01 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Molecular basis for the functions of dominantly active Y35N and inactive D60K Rheb mutants in mTORC1 signaling.
J Mol Cell Biol, 12, 2020
|
|
