5JON
 
 | | Crystal structure of the unliganded form of HCN2 CNBD | | Descriptor: | Maltose-binding periplasmic protein,Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2, NITRATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Klenchin, V.A, Chanda, B. | | Deposit date: | 2016-05-02 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Structure and dynamics underlying elementary ligand binding events in human pacemaking channels.
Elife, 5, 2016
|
|
1Z56
 
 | | Co-Crystal Structure of Lif1p-Lig4p | | Descriptor: | DNA ligase IV, Ligase interacting factor 1 | | Authors: | Dore, A.S, Furnham, N, Davies, O.R, Sibanda, B.L, Chirgadze, D.Y, Jackson, S.P, Pellegrini, L, Blundell, T.L. | | Deposit date: | 2005-03-17 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode.
DNA REPAIR, 5, 2006
|
|
3FL2
 
 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
6LOL
 
 | | The crystal structure of full length IpaH9.8 | | Descriptor: | E3 ubiquitin-protein ligase ipaH9.8 | | Authors: | Ye, Y, Huang, H. | | Deposit date: | 2020-01-06 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
2QS4
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | Descriptor: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS1
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS2
 
 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | Descriptor: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Alushin, G.M, Jane, D.E, Mayer, M.L. | | Deposit date: | 2007-07-30 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
3BA8
 
 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 2-amino-7-fluoro-5-oxo-5H-chromeno[2,3-b]pyridine-3-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAB
 
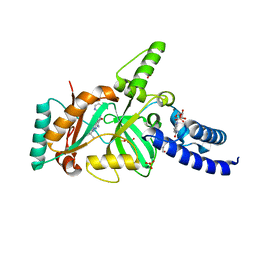 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-(4-pyrimidin-2-ylpiperazin-1-yl)pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
3BAA
 
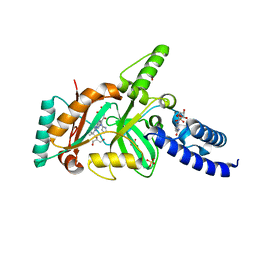 | | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase | | Descriptor: | 7-amino-2-tert-butyl-4-{[2-(1H-imidazol-4-yl)ethyl]amino}pyrido[2,3-d]pyrimidine-6-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Inhibition of Bacterial NAD+ Dependent DNA Ligase
To be Published
|
|
2JJN
 
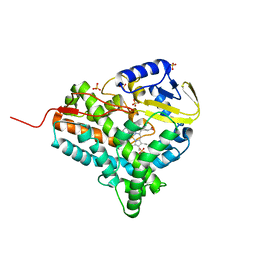 | | Structure of closed cytochrome P450 EryK | | Descriptor: | CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JJO
 
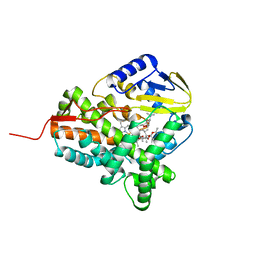 | | Structure of cytochrome P450 EryK in complex with its natural substrate erD | | Descriptor: | CYTOCHROME P450 113A1, Erythromycin D, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
8HAE
 
 | | Cryo-EM structure of HACE1 dimer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J, Machida, S. | | Deposit date: | 2022-10-26 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8H8X
 
 | | Cryo-EM structure of HACE1 monomer | | Descriptor: | E3 ubiquitin-protein ligase HACE1 | | Authors: | Singh, S, Machida, S, Tulsian, N.K, Choong, Y.K, Ng, J, Shanker, S, Yaochen, L.D, Shi, J, Sivaraman, J. | | Deposit date: | 2022-10-24 | | Release date: | 2023-06-28 | | Last modified: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Structural Basis for the Enzymatic Activity of the HACE1 HECT-Type E3 Ligase Through N-Terminal Helix Dimerization.
Adv Sci, 10, 2023
|
|
8G7U
 
 | |
8G7T
 
 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-end) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G7V
 
 | | Cryo-EM structure of Riplet:RIG-I:dsRNA complex (end-inter) | | Descriptor: | Antiviral innate immune response receptor RIG-I, E3 ubiquitin-protein ligase RNF135, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-11-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The E3 ligase Riplet promotes RIG-I signaling independent of RIG-I oligomerization.
Nat Commun, 14, 2023
|
|
8G4Y
 
 | | Structure of ZNRF3 ECD bound to peptide MK1-3.6.10 | | Descriptor: | E3 ubiquitin-protein ligase ZNRF3, MK1-3.6.10 | | Authors: | Harris, S.F, Wu, P. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Potent and selective binders of the E3 ubiquitin ligase ZNRF3 stimulate Wnt signaling and intestinal organoid growth.
Cell Chem Biol, 31, 2024
|
|
4FNA
 
 | | Structure of unliganded FhuD2 from Staphylococcus Aureus | | Descriptor: | Ferric hydroxamate receptor 2, SULFATE ION | | Authors: | Shilton, B.H, Heinrichs, D.E. | | Deposit date: | 2012-06-19 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal and solution structure analysis of FhuD2 from Staphylococcus aureus in multiple unliganded conformations and bound to ferrioxamine-B.
Biochemistry, 53, 2014
|
|
4FKM
 
 | |
3BL3
 
 | | tRNA guanine transglycosylase V233G mutant apo structure | | Descriptor: | GLYCEROL, Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Tidten, N, Heine, A, Reuter, K, Klebe, G. | | Deposit date: | 2007-12-10 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Investigation of Specificity Determinants in Bacterial tRNA-Guanine Transglycosylase Reveals Queuine, the Substrate of Its Eucaryotic Counterpart, as Inhibitor
Plos One, 8, 2013
|
|
4UDD
 
 | | GR in complex with desisobutyrylciclesonide | | Descriptor: | 1,2-ETHANEDIOL, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DESISOBUYTYRYL CICLESONIDE, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDC
 
 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UDB
 
 | | MR in complex with desisobutyrylciclesonide | | Descriptor: | DESISOBUYTYRYL CICLESONIDE, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, JellesmarkJensen, T, Cavallin, A, Nilsson, E, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
3BA9
 
 | | Structural Basis for Inhbition of NAD-Dependent Ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, GLYCEROL, ... | | Authors: | Pinko, C. | | Deposit date: | 2007-11-07 | | Release date: | 2008-11-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for Inhbition of NAD-Dependent Ligase
To be Published
|
|
