6XBW
 
 | | Cryo-EM structure of V-ATPase from bovine brain, state 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
5L5J
 
 | |
7DSV
 
 | | Structure of a human NHE1-CHP1 complex under pH 6.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Calcineurin B homologous protein 1, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
2MJQ
 
 | |
5L62
 
 | | Yeast 20S proteasome with human beta5c (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 16 | | Descriptor: | (2~{S})-3-(1~{H}-indol-3-yl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6XZA
 
 | | E. coli 70S ribosome in complex with dirithromycin, and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
7DSW
 
 | | Structure of a human NHE1-CHP1 complex under pH 7.5 | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Sodium/hydrogen exchanger 1 | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
8AVQ
 
 | | AO75L in Complex with UDP-Xylose | | Descriptor: | 1,2-ETHANEDIOL, AO75L, BICINE, ... | | Authors: | Laugeri, M.E, Speciale, I, Gimeno, A, Lin, S, Poveda, A, Lowary, T, Van Etten, J.L, Barbero, J.J, De Castro, C, Tonetti, M, Rojas, A.L. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | AO75L in Complex with UDP-Xylose
To Be Published
|
|
6XHV
 
 | | Crystal structure of the A2058-dimethylated Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A- and P-site tRNAs, and deacylated E-site tRNA at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svetlov, M.S, Syroegin, E.A, Aleksandrova, E.V, Atkinson, G.C, Gregory, S.T, Mankin, A.S, Polikanov, Y.S. | | Deposit date: | 2020-06-19 | | Release date: | 2020-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance.
Nat.Chem.Biol., 17, 2021
|
|
6VZJ
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing mRNA with a 15 nt long spacer, fMet-tRNAs at E-site and P-site, and lacking transcription factor NusG | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6XRV
 
 | |
7DSX
 
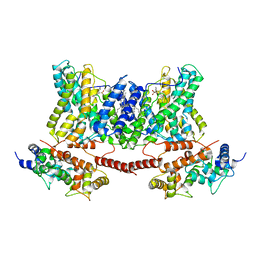 | | Structure of a human NHE1-CHP1 complex under pH 7.5, bound by cariporide | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, Calcineurin B homologous protein 1, N-[bis(azanyl)methylidene]-3-methylsulfonyl-4-propan-2-yl-benzamide, ... | | Authors: | Dong, Y, Gao, Y, Li, B, Zhang, X.C, Zhao, Y. | | Deposit date: | 2021-01-03 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and mechanism of the human NHE1-CHP1 complex.
Nat Commun, 12, 2021
|
|
6X9N
 
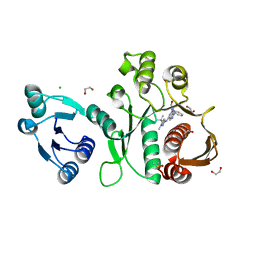 | | Pseudomonas aeruginosa MurC with AZ5595 | | Descriptor: | (2R)-2-({4-[(5-tert-butyl-1-methyl-1H-pyrazol-3-yl)amino]-1H-pyrazolo[3,4-d]pyrimidin-6-yl}amino)-2-phenylethan-1-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Horanyi, P.S, Mayclin, S.J, Durand-Reville, T.F, Lorimer, D.D, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Pseudomonas aeruginosa MurC with AZ5595
To Be Published
|
|
6W2S
 
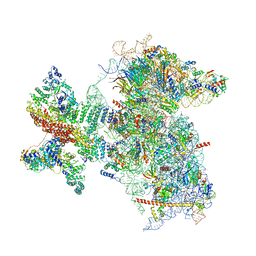 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
8X41
 
 | | Crystal structure of DIMT1 in complex with 5'-methylthioadenosine and adenosine from Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, ACETATE ION, ... | | Authors: | Saha, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
7DDQ
 
 | | Structure of RC-LH1-PufX from Rhodobacter veldkampii | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Bracun, L, Yamagata, A, Shirouzu, M, Liu, L.N. | | Deposit date: | 2020-10-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM structure of the photosynthetic RC-LH1-PufX supercomplex at 2.8-angstrom resolution.
Sci Adv, 7, 2021
|
|
5L52
 
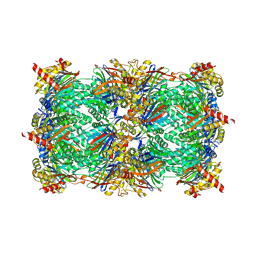 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 14 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
5L5S
 
 | |
6XBY
 
 | | Cryo-EM structure of V-ATPase from bovine brain, state 2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Wang, R, Li, X. | | Deposit date: | 2020-06-07 | | Release date: | 2020-08-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures of intact V-ATPase from bovine brain.
Nat Commun, 11, 2020
|
|
6XF5
 
 | | Cryo-EM structure of a biotinylated SARS-CoV-2 spike probe in the prefusion state (RBDs down) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Cerutti, G, Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-06-15 | | Release date: | 2020-09-02 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structure-Based Design with Tag-Based Purification and In-Process Biotinylation Enable Streamlined Development of SARS-CoV-2 Spike Molecular Probes.
SSRN, 2020
|
|
8WHT
 
 | |
5L6A
 
 | | Yeast 20S proteasome with mouse beta5i (1-138) and mouse beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | (2~{S})-3-(4-methoxyphenyl)-~{N}-[(2~{S},3~{S},4~{R})-4-methyl-3,5-bis(oxidanyl)-1-phenyl-pentan-2-yl]-2-[[(2~{R})-2-(2-morpholin-4-ylethanoylamino)propanoyl]amino]propanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
8AXG
 
 | | Crystal structure of Fusobacterium nucleatum fusolisin protease | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Fusolisin, ... | | Authors: | Isupov, M.N, Wiener, R, Rouvinski, A, Fahoum, J, Kumar, M, Read, R.J. | | Deposit date: | 2022-08-31 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure of Fusobacterium nucleatum fusolisin protease
To Be Published
|
|
5LAI
 
 | | Ligand-induced aziridine-formation at the yeast proteasomal subunit beta5 by sulfonate esters | | Descriptor: | (2S,3S)-3-methylaziridine-2-carboxylic acid, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Groll, M, Dubiella, C, Cui, H. | | Deposit date: | 2016-06-14 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Tunable Probes with Direct Fluorescence Signals for the Constitutive and Immunoproteasome.
Angew. Chem. Int. Ed. Engl., 55, 2016
|
|
8X3W
 
 | | Crystal structure of DIMT1 from the thermophilic archaeon, Pyrococcus horikoshii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Sayan, S, Mandal, S.K, Dutta, A, Kanaujia, S.P. | | Deposit date: | 2023-11-14 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural and functional characterization of archaeal DIMT1 unveils distinct protein dynamics essential for efficient catalysis.
Structure, 32, 2024
|
|
