5EYT
 
 | | Crystal Structure of Adenylosuccinate Lyase from Schistosoma mansoni in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Adenylosuccinate lyase | | Authors: | Romanello, L, Torini, J.R, Bird, L, Nettleship, J, Owens, R, Reddivari, Y, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2015-11-25 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3649 Å) | | Cite: | Structural and kinetic analysis of Schistosoma mansoni Adenylosuccinate Lyase (SmADSL).
Mol. Biochem. Parasitol., 214, 2017
|
|
3NGT
 
 | | Structure of Leishmania NDKb complexed with AMP. | | Descriptor: | ADENOSINE MONOPHOSPHATE, Nucleoside diphosphate kinase | | Authors: | Trindade, D.M, Sousa, T.A.C.B, Tonoli, C.C.C, Santos, C.R, Arni, R.K, Ward, R.J, Oliveira, A.H.C, Murakami, M.T. | | Deposit date: | 2010-06-13 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Molecular adaptability of nucleoside diphosphate kinase b from trypanosomatid parasites: stability, oligomerization and structural determinants of nucleotide binding.
Mol Biosyst, 7, 2011
|
|
5XET
 
 | | Crystal structure of Mycobacterium tuberculosis methionyl-tRNA synthetase bound by methionyl-adenylate (Met-AMP) | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methionine--tRNA ligase, ... | | Authors: | Wang, W, Qin, B, Wojdyla, J.A, Wang, M, Gao, X, Cui, S. | | Deposit date: | 2017-04-06 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural characterization of free-state and product-stateMycobacterium tuberculosismethionyl-tRNA synthetase reveals an induced-fit ligand-recognition mechanism.
IUCrJ, 5, 2018
|
|
2VFL
 
 | | AKAP18 delta central domain - CMP | | Descriptor: | AKAP18 DELTA, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Gold, M.G, Smith, F.D, Scott, J.D, Barford, D. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-06 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Akap18 Contains a Phosphoesterase Domain that Binds AMP
J.Mol.Biol., 375, 2008
|
|
5GTD
 
 | |
6F2V
 
 | | Crystal structure of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3) in complex with AMP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Dohler, C, Zebisch, M, Strater, N. | | Deposit date: | 2017-11-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and substrate binding mode of ectonucleotide phosphodiesterase/pyrophosphatase-3 (NPP3).
Sci Rep, 8, 2018
|
|
1V8S
 
 | | Crystal structure analusis of the ADP-ribose pyrophosphatase complexed with AMP and Mg | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADP-ribose pyrophosphatase, MAGNESIUM ION | | Authors: | Yoshiba, S, Ooga, T, Nakagawa, N, Shibata, T, Inoue, Y, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-14 | | Release date: | 2004-10-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into the Thermus thermophilus ADP-ribose pyrophosphatase mechanism via crystal structures with the bound substrate and metal
J.Biol.Chem., 279, 2004
|
|
5HM3
 
 | | 2.25 Angstrom Resolution Crystal Structure of Long-chain-fatty-acid-AMP Ligase FadD32 from Mycobacterium tuberculosis in complex with Inhibitor 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine | | Descriptor: | 5'-O-[(11-phenoxyundecanoyl)sulfamoyl]adenosine, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Minasov, G, Warwrzak, Z, Kuhn, M.L, Shuvalova, L, Flores, K.J, Wilson, D.J, Grimes, K.D, Aldrich, C.C, Anderson, W.A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Essential Mtb FadD32 Enzyme: A Promising Drug Target for Treating Tuberculosis.
Acs Infect Dis., 2, 2016
|
|
3SZG
 
 | | Crystal structure of C176A glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi and NaAD+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, Glutamine-dependent NAD(+) synthetase, ... | | Authors: | Chuenchor, W, Doukov, T, Gerratana, B. | | Deposit date: | 2011-07-19 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
3SYT
 
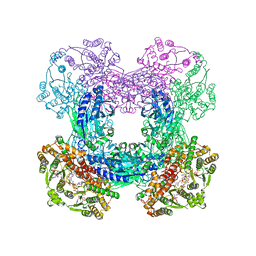 | | Crystal structure of glutamine-dependent NAD+ synthetase from M. tuberculosis bound to AMP/PPi, NAD+, and glutamate | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Chuenchor, W, Gerratana, B. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6511 Å) | | Cite: | Regulation of the intersubunit ammonia tunnel in Mycobacterium tuberculosis glutamine-dependent NAD+ synthetase.
Biochem.J., 443, 2012
|
|
5IKP
 
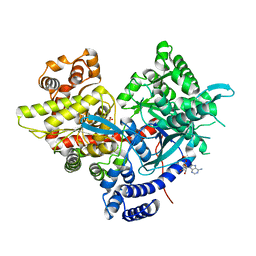 | | Crystal structure of human brain glycogen phosphorylase bound to AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycogen phosphorylase, brain form, ... | | Authors: | Mathieu, C, Li de la Sierra-Gallay, I, Xu, X, Haouz, A, Rodrigues-Lima, F. | | Deposit date: | 2016-03-03 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights into Brain Glycogen Metabolism: THE STRUCTURE OF HUMAN BRAIN GLYCOGEN PHOSPHORYLASE.
J.Biol.Chem., 291, 2016
|
|
3IB7
 
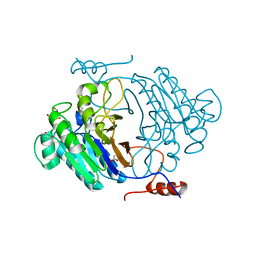 | | Crystal structure of full length Rv0805 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Podobnik, M, Dermol, U. | | Deposit date: | 2009-07-15 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A mycobacterial cyclic AMP phosphodiesterase that moonlights as a modifier of cell wall permeability
J.Biol.Chem., 284, 2009
|
|
1J1Z
 
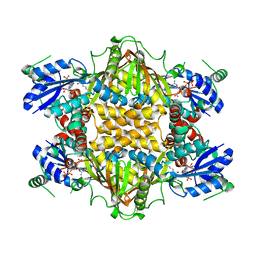 | |
3TY9
 
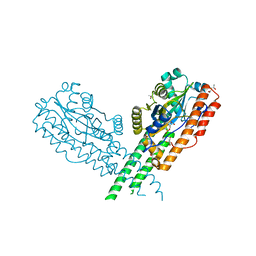 | | Crystal Structure of C. Thermocellum PNKP Ligase Domain AMP-Adenylate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Smith, P, Wang, L, Shuman, S. | | Deposit date: | 2011-09-24 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | The adenylyltransferase domain of bacterial Pnkp defines a unique RNA ligase family.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2E41
 
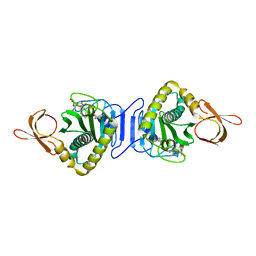 | | Crystal Structure Of Biotin Protein Ligase From Pyrococcus Horikoshii Complexed with the Reaction Product Analog Biotinol-5'-AMP, Mutations R48A and K111A | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, biotin--[acetyl-CoA-carboxylase] ligase | | Authors: | Bagautdinov, B, Matsuura, Y, Bagautdinova, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-01 | | Release date: | 2007-06-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Protein biotinylation visualized by a complex structure of biotin protein ligase with a substrate
J.Biol.Chem., 283, 2008
|
|
1ECJ
 
 | |
2E69
 
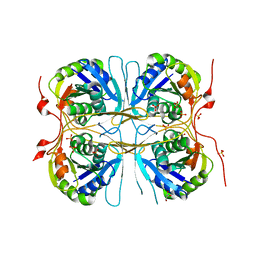 | |
2E6E
 
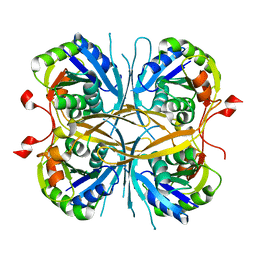 | |
3A7A
 
 | | Crystal structure of E. coli lipoate-protein ligase A in complex with octyl-amp and apoH-protein | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glycine cleavage system H protein, Lipoate-protein ligase A, ... | | Authors: | Fujiwara, K, Hosaka, H, Nakagawa, A. | | Deposit date: | 2009-09-20 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Global conformational change associated with the two-step reaction catalyzed by Escherichia coli lipoate-protein ligase A.
J.Biol.Chem., 285, 2010
|
|
3A7R
 
 | |
5CK7
 
 | |
2E6G
 
 | |
2E6B
 
 | |
5WS9
 
 | | Pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, ATP and allosteric activator AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
1YXU
 
 | | Crystal Structure of Kinase Pim1 in Complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Kumar, A, Mandiyan, V, Suzuki, Y, Zhang, C, Rice, J, Tsai, J, Artis, D.R, Ibrahim, P, Bremer, R. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structures of Proto-oncogene Kinase Pim1: A Target of Aberrant Somatic Hypermutations in Diffuse Large Cell Lymphoma.
J.Mol.Biol., 348, 2005
|
|
