2A47
 
 | | Crystal structure of amFP486 H199T | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A48
 
 | | Crystal structure of amFP486 E150Q | | Descriptor: | BETA-MERCAPTOETHANOL, GFP-like fluorescent chromoprotein amFP486 | | Authors: | Henderson, J.N, Remington, S.J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and mutational analysis of amFP486, a cyan fluorescent protein from Anemonia majano
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A49
 
 | | Crystal structure of clavulanic acid bound to E166A variant of SHV-1 beta-lactamase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | Padayatti, P.S, Helfand, M.S, Totir, M.A, Carey, M.P, Carey, P.R, Bonomo, R.A, van den Akker, F. | | Deposit date: | 2005-06-28 | | Release date: | 2005-08-02 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | High Resolution Crystal Structures of the trans-Enamine Intermediates Formed by Sulbactam and Clavulanic Acid and E166A SHV-1 {beta}-Lactamase.
J.Biol.Chem., 280, 2005
|
|
2A4A
 
 | | Deoxyribose-phosphate aldolase from P. yoelii | | Descriptor: | deoxyribose-phosphate aldolase | | Authors: | Walker, J.R, Amani, M, Lew, J, Wiegelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
2A4C
 
 | | Crystal structure of mouse cadherin-11 EC1 | | Descriptor: | Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Schieren, I, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A4D
 
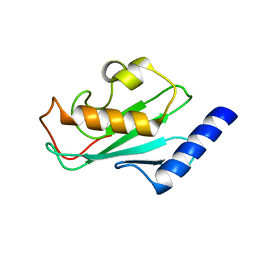 | | Structure of the human ubiquitin-conjugating enzyme E2 variant 1 (UEV-1) | | Descriptor: | Ubiquitin-conjugating enzyme E2 variant 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Mackenzie, F, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-06-28 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen.
Mol Cell Proteomics, 11, 2012
|
|
2A4E
 
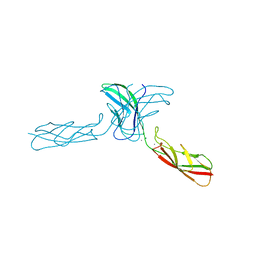 | | Crystal structure of mouse cadherin-11 EC1-2 | | Descriptor: | CALCIUM ION, Cadherin-11 | | Authors: | Patel, S.D, Ciatto, C, Chen, C.P, Bahna, F, Arkus, N, Rajebhosale, M, Jessell, T.M, Honig, B, Price, S.R, Shapiro, L. | | Deposit date: | 2005-06-28 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Type II cadherin ectodomain structures: implications for classical cadherin specificity.
Cell(Cambridge,Mass.), 124, 2006
|
|
2A4F
 
 | | Synthesis and Activity of N-Axyl Azacyclic Urea HIV-1 Protease Inhibitors with High Potency Against Multiple Drug Resistant Viral Strains. | | Descriptor: | (5R,6R)-5-BENZYL-6-HYDROXY-2,4-BIS(4-HYDROXY-3-METHOXYBENZYL)-1-[3-(4-HYDROXYPHENYL)PROPANOYL]-1,2,4-TRIAZEPAN-3-ONE, Pol polyprotein | | Authors: | Zhao, C, Sham, H, Sun, M, Lin, S, Stoll, V, Stewart, K.D, Mo, H, Vasavanonda, S, Saldivar, A, McDonald, E. | | Deposit date: | 2005-06-28 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4G
 
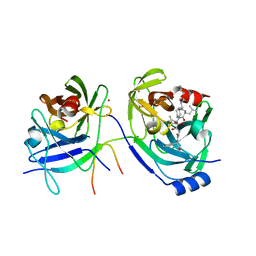 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4H
 
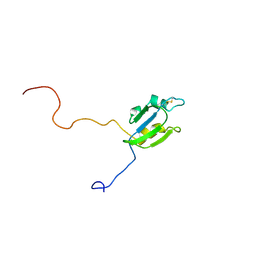 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
2A4J
 
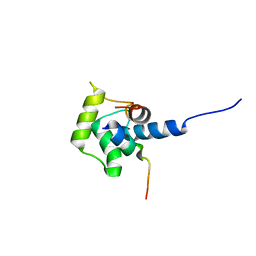 | | Solution structure of the C-terminal domain (T94-Y172) of the human centrin 2 in complex with a 17 residues peptide (P1-XPC) from xeroderma pigmentosum group C protein | | Descriptor: | 17-mer peptide P1-XPC from DNA-repair protein complementing XP-C cells, Centrin 2 | | Authors: | Yang, A, Miron, S, Mouawad, L, Duchambon, P, Blouquit, Y, Craescu, C.T. | | Deposit date: | 2005-06-29 | | Release date: | 2005-07-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Flexibility and plasticity of human centrin 2 binding to the xeroderma pigmentosum group C protein (XPC) from nuclear excision repair.
Biochemistry, 45, 2006
|
|
2A4K
 
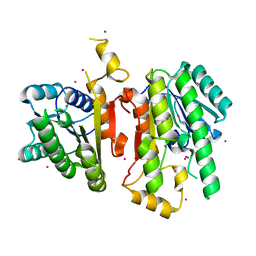 | | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137 | | Descriptor: | 3-oxoacyl-[acyl carrier protein] reductase, UNKNOWN ATOM OR ION | | Authors: | Zhou, W, Ebihara, A, Tempel, W, Yokoyama, S, Chen, L, Kuramitsu, S, Nguyen, J, Chang, S.-H, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-29 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | 3-Oxoacyl-[acyl carrier protein] reductase from Thermus thermophilus TT0137
To be Published
|
|
2A4L
 
 | |
2A4M
 
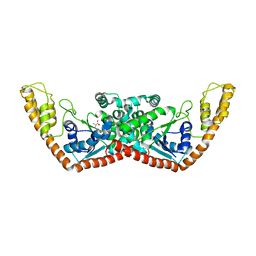 | | Structure of Trprs II bound to ATP | | Descriptor: | TRYPTOPHAN, Tryptophanyl-tRNA synthetase II | | Authors: | Buddha, M.R, Crane, B.R. | | Deposit date: | 2005-06-29 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Tryptophanyl-tRNA Synthetase II from Deinococcus radiodurans Bound to ATP and Tryptophan: Insight into subunit cooperativity and domain motions linked to catalysis
J.Biol.Chem., 280, 2005
|
|
2A4N
 
 | | Crystal structure of aminoglycoside 6'-N-acetyltransferase complexed with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Xiong, B, Breitbach, C, Berghuis, A.M. | | Deposit date: | 2005-06-29 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of aminoglycoside acetyltransferase AAC(6')-Ii in a novel crystal form: structural and normal-mode analyses.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2A4O
 
 | | Dual modes of modification of Hepatitis A virus 3C protease by a serine derived beta-lactone: selective crytstallization and high resolution structure of the His102 adduct | | Descriptor: | ACETYL GROUP, N-[(BENZYLOXY)CARBONYL]-L-ALANINE, PHENYLALANINE AMIDE, ... | | Authors: | Yin, J, Bergmann, E.M, Cherney, M.M, Lall, M.S, Jain, R.P, Vederas, J.C, James, M.N.G. | | Deposit date: | 2005-06-29 | | Release date: | 2005-12-27 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Dual Modes of Modification of Hepatitis A Virus 3C Protease by a Serine-derived beta-Lactone: Selective Crystallization and Formation of a Functional Catalytic Triad in the Active Site
J.MOL.BIOL., 354, 2005
|
|
2A4Q
 
 | | HCV NS3 protease with NS4a peptide and a covalently bound macrocyclic ketoamide compound. | | Descriptor: | (2R)-({N-[(3S)-3-({[(3S,6S)-6-CYCLOHEXYL-5,8-DIOXO-4,7-DIAZABICYCLO[14.3.1]ICOSA-1(20),16,18-TRIEN-3-YL]CARBONYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETIC ACID, BETA-MERCAPTOETHANOL, NS3 protease/helicase', ... | | Authors: | Chen, K.X, Njoroge, F.G, Prongay, A, Pichardo, J, Madison, V, Girijavallabhan, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Synthesis and Biological Activity of Macrocyclic Inhibitors of Hepatitis C Virus (HCV) NS3 Protease
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4R
 
 | | HCV NS3 Protease Domain with a Ketoamide Inhibitor Covalently bound. | | Descriptor: | NS3 protease/helicase, Ns4a peptide, ZINC ION, ... | | Authors: | Bogen, S, Saksena, A.K, Arasappan, A, Gu, H, Njoroge, F.G, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Prongay, A, Madison, V. | | Deposit date: | 2005-06-29 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Hepatitis C Virus NS3-4A serine protease inhibitors: Use of a P2-P1 cyclopropyl alanine combination for improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
2A4T
 
 | | Crystal structure of spin labeled T4 Lysozyme (V131R7) | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, CHLORIDE ION, ... | | Authors: | Fleissner, M.R, Cascio, D, Sawaya, M.R, Hideg, K, Hubbell, W.L. | | Deposit date: | 2005-06-29 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of spin labeled T4 Lysozyme (V131R7
To be Published
|
|
2A4V
 
 | | Crystal Structure of a truncated mutant of yeast nuclear thiol peroxidase | | Descriptor: | Peroxiredoxin DOT5 | | Authors: | Choi, J, Choi, S, Chon, J.-K, Choi, J, Cha, M.-K, Kim, I.-H, Shin, W. | | Deposit date: | 2005-06-29 | | Release date: | 2006-03-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the C107S/C112S mutant of yeast nuclear 2-Cys peroxiredoxin
Proteins, 61, 2005
|
|
2A4W
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Copper(II)-Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, COPPER (II) ION, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
2A4X
 
 | | Crystal Structure Of Mitomycin C-Binding Protein Complexed with Metal-Free Bleomycin A2 | | Descriptor: | BLEOMYCIN A2, Mitomycin-Binding Protein | | Authors: | Danshiitsoodol, N, de Pinho, C.A, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2005-06-30 | | Release date: | 2006-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Mitomycin C (MMC)-binding Protein from MMC-producing Microorganisms Protects from the Lethal Effect of Bleomycin: Crystallographic Analysis to Elucidate the Binding Mode of the Antibiotic to the Protein
J.Mol.Biol., 360, 2006
|
|
2A4Z
 
 | | Crystal Structure of human PI3Kgamma complexed with AS604850 | | Descriptor: | (5E)-5-[(2,2-DIFLUORO-1,3-BENZODIOXOL-5-YL)METHYLENE]-1,3-THIAZOLIDINE-2,4-DIONE, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit, gamma isoform | | Authors: | Camps, M, Ruckle, T, Ji, H, Ardissone, V, Rintelen, F, Shaw, J, Ferrandi, C, Chabert, C, Gillieron, C, Francon, B, Martin, T, Gretener, D, Perrin, D, Leroy, D, Vitte, P.-A, Hirsch, E, Wymann, M.P, Cirillo, R, Schwarz, M.K, Rommel, C. | | Deposit date: | 2005-06-30 | | Release date: | 2005-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Blockade of PI3Kgamma suppresses joint inflammation and damage in mouse models of rheumatoid arthritis
NAT.MED. (N.Y.), 11, 2005
|
|
2A50
 
 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A51
 
 | | Structure of the (13-51) domain of the nucleocapsid protein NCp8 from SIVlhoest | | Descriptor: | ZINC ION, nucleocapsid protein | | Authors: | Morellet, N, Meudal, H, Bouaziz, S, Roques, B.P. | | Deposit date: | 2005-06-30 | | Release date: | 2006-06-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the zinc finger domain encompassing residues 13-51 of the nucleocapsid protein from simian immunodeficiency virus
Biochem.J., 393, 2006
|
|
