3E5N
 
 | | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331 | | Descriptor: | D-alanine-D-alanine ligase A | | Authors: | Doan, T.N.T, Kim, J.K, Kim, H.S, Ahn, Y.J, Kim, J.G, Lee, B.M, Kang, L.W. | | Deposit date: | 2008-08-14 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of D-alanine-D-alanine ligase from Xanthomonas oryzae pv. oryzae KACC10331
To be published
|
|
1YCA
 
 | | DISTAL POCKET POLARITY IN LIGAND BINDING TO MYOGLOBIN: DEOXY AND CARBONMONOXY FORMS OF A THREONINE68 (E11) MUTANT INVESTIGATED BY X-RAY CRYSTALLOGRAPHY AND INFRARED SPECTROSCOPY | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Cameron, A.D, Smerdon, S.J, Wilkinson, A.J, Habash, J, Helliwell, J.R. | | Deposit date: | 1993-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Distal pocket polarity in ligand binding to myoglobin: deoxy and carbonmonoxy forms of a threonine68(E11) mutant investigated by X-ray crystallography and infrared spectroscopy.
Biochemistry, 32, 1993
|
|
1ZNG
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, HEPTAN-1-OL, Major Urinary Protein | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
6CU0
 
 | | Crystal structure of 4-1BBL/4-1BB (C121S) complex in P21 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Tumor necrosis factor ligand superfamily member 9, Tumor necrosis factor receptor superfamily member 9 | | Authors: | Aruna, B, Zajonc, D.M, Doukov, T. | | Deposit date: | 2018-03-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
3KO9
 
 | |
1ZND
 
 | | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex | | Descriptor: | CADMIUM ION, Major Urinary Protein, PENTAN-1-OL | | Authors: | Malham, R, Johnstone, S, Bingham, R.J, Barratt, E, Phillips, S.E, Laughton, C.A, Homans, S.W. | | Deposit date: | 2005-05-11 | | Release date: | 2005-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Strong Solute-Solute Dispersive Interactions in a Protein-Ligand Complex.
J.Am.Chem.Soc., 127, 2005
|
|
4NKH
 
 | | Crystal structure of SspH1 LRR domain | | Descriptor: | E3 ubiquitin-protein ligase sspH1 | | Authors: | Keszei, A.F.A, Xiaojing, T, Mccormick, C, Zeqiraj, E, Rohde, J.R, Tyers, M, Sicheri, F. | | Deposit date: | 2013-11-12 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of an SspH1-PKN1 Complex Reveals the Basis for Host Substrate Recognition and Mechanism of Activation for a Bacterial E3 Ubiquitin Ligase.
Mol.Cell.Biol., 34, 2014
|
|
7U35
 
 | |
4S3O
 
 | | PCGF5-RING1B-UbcH5c complex | | Descriptor: | E3 ubiquitin-protein ligase RING2, Polycomb group RING finger protein 5, Ubiquitin-conjugating enzyme E2 D3, ... | | Authors: | Taherbhoy, A.M, Cochran, A.G. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | BMI1-RING1B is an autoinhibited RING E3 ubiquitin ligase.
Nat Commun, 6, 2015
|
|
7MWE
 
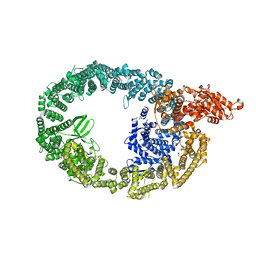 | | HUWE1 in map with focus on WWE | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
3KOB
 
 | |
3KOC
 
 | |
7MWD
 
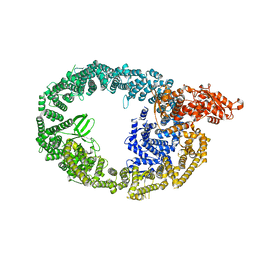 | | HUWE1 in map with focus on HECT | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
3FL2
 
 | | Crystal structure of the ring domain of the E3 ubiquitin-protein ligase UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-12-18 | | Release date: | 2009-01-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of the Ring Domain of the E3 Ubiquitin-Protein Ligase Uhrf1
To be Published
|
|
7MWF
 
 | | HUWE1 in map with focus on interface | | Descriptor: | E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
7MOP
 
 | | Cryo-EM structure of human HUWE1 in complex with DDIT4 | | Descriptor: | DNA damage-inducible transcript 4 protein, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Hunkeler, M, Fischer, E.S. | | Deposit date: | 2021-05-03 | | Release date: | 2021-07-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Solenoid architecture of HUWE1 contributes to ligase activity and substrate recognition.
Mol.Cell, 81, 2021
|
|
8IUM
 
 | | Cryo-EM structure of the tafluprost acid-bound human PTGFR-Gq complex | | Descriptor: | (~{Z})-7-[(1~{R},2~{R},3~{R},5~{S})-2-[(~{E})-3,3-bis(fluoranyl)-4-phenoxy-but-1-enyl]-3,5-bis(oxidanyl)cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUK
 
 | | Cryo-EM structure of the PGF2-alpha-bound human PTGFR-Gq complex | | Descriptor: | (Z)-7-[(1R,2R,3R,5S)-3,5-bis(oxidanyl)-2-[(E,3S)-3-oxidanyloct-1-enyl]cyclopentyl]hept-5-enoic acid, Antibody fragment scFv16, G subunit alpha (q), ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
8IUL
 
 | | Cryo-EM structure of the latanoprost-bound human PTGFR-Gq complex | | Descriptor: | Antibody fragment scFv16, G subunit alpha (q), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wu, C, Xu, Y, Xu, H.E. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Ligand-induced activation and G protein coupling of prostaglandin F 2 alpha receptor.
Nat Commun, 14, 2023
|
|
3KNF
 
 | |
3QT0
 
 | | Revealing a steroid receptor ligand as a unique PPARgamma agonist | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, Nuclear receptor coactivator 1 peptide, Peroxisome proliferator-activated receptor gamma | | Authors: | Rong, H. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.496 Å) | | Cite: | Revealing a steroid receptor ligand as a unique PPAR gamma agonist.
Cell Res., 22, 2012
|
|
1FOT
 
 | |
6CPR
 
 | | Crystal structure of 4-1BBL/4-1BB complex in C2 space group | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Aruna, B, Zajonc, D.M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-05-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of the human 4-1BB receptor bound to its ligand 4-1BBL reveal covalent receptor dimerization as a potential signaling amplifier.
J. Biol. Chem., 293, 2018
|
|
1JWN
 
 | | Crystal Structure of Scapharca inaequivalvis HbI, I114F Mutant Ligated to Carbon Monoxide. | | Descriptor: | CARBON MONOXIDE, Globin I - Ark Shell, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Gibson, Q.H, Cushing, L, Royer Jr, W.E. | | Deposit date: | 2001-09-04 | | Release date: | 2001-12-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Restricting the ligand-linked heme movement in Scapharca dimeric hemoglobin reveals tight coupling between distal and proximal contributions to cooperativity.
Biochemistry, 40, 2001
|
|
