7KPV
 
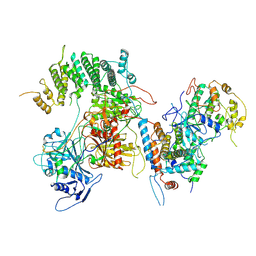 | | Structure of kinase and Central lobes of yeast CKM | | Descriptor: | Mediator of RNA polymerase II transcription subunit 12, Mediator of RNA polymerase II transcription subunit 13, Meiotic mRNA stability protein kinase SSN3, ... | | Authors: | Li, Y.C, Chao, T.C, Kim, H.J, Cholko, T, Chen, S.F, Nakanishi, K, Chang, C.E, Murakami, K, Garcia, B.A, Boyer, T.G, Tsai, K.L. | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and noncanonical Cdk8 activation mechanism within an Argonaute-containing Mediator kinase module.
Sci Adv, 7, 2021
|
|
7KXE
 
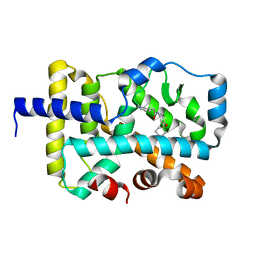 | |
7KQJ
 
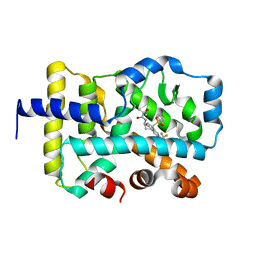 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH A NOVEL TRICYCLIC-CARBOCYLIC RORGT INVERSE AGONIST | | Descriptor: | N-[(3R,3aS,9bS)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[a]naphthalen-3-yl]-2-hydroxy-2-methylpropanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Tricyclic-Carbocyclic ROR gamma t Inverse Agonists-Discovery of BMS-986313.
J.Med.Chem., 64, 2021
|
|
7KXF
 
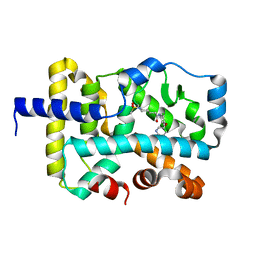 | |
3J7Q
 
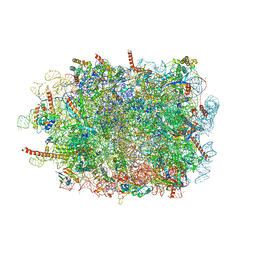 | | Structure of the idle mammalian ribosome-Sec61 complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Voorhees, R.M, Fernandez, I.S, Scheres, S.H.W, Hegde, R.S. | | Deposit date: | 2014-08-01 | | Release date: | 2014-09-03 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Mammalian ribosome-sec61 complex to 3.4 a resolution.
Cell(Cambridge,Mass.), 157, 2014
|
|
1LFP
 
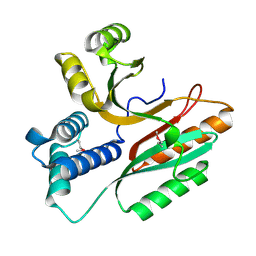 | | Crystal Structure of a Conserved Hypothetical Protein Aq1575 from Aquifex Aeolicus | | Descriptor: | Hypothetical protein AQ_1575 | | Authors: | Shin, D.H, Yokota, H, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2002-04-11 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of conserved hypothetical protein Aq1575 from Aquifex aeolicus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2N54
 
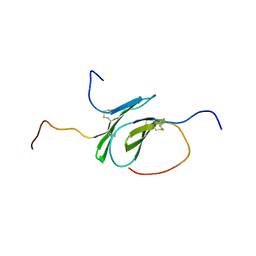 | | Solution structure of a disulfide stabilized XCL1 dimer | | Descriptor: | Lymphotactin | | Authors: | Tyler, R.C, Tuinstra, R.L, Peterson, F.F, Volkman, B.F. | | Deposit date: | 2015-07-07 | | Release date: | 2015-10-07 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Engineering Metamorphic Chemokine Lymphotactin/XCL1 into the GAG-Binding, HIV-Inhibitory Dimer Conformation.
Acs Chem.Biol., 10, 2015
|
|
4NYX
 
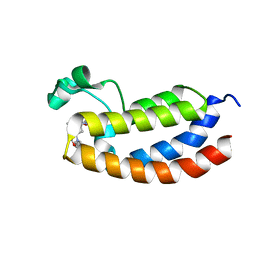 | | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand | | Descriptor: | (3R)-N-[3-(7-methoxy-3,4-dihydroquinolin-1(2H)-yl)propyl]-3-methyl-2-oxo-1,2,3,4-tetrahydroquinoxaline-5-carboxamide, CREB-binding protein | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Rooney, T.P.C, Fedorov, O, Martin, S, Monteiro, O.P, Conway, S.J, von Delft, F, Brennan, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-11 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal Structure of the Bromodomain of human CREBBP in complex with a dihydroquinoxalinone ligand
TO BE PUBLISHED
|
|
1NQ0
 
 | | TR Receptor Mutations Conferring Hormone Resistance and Reduced Corepressor Release Exhibit Decreased Stability in the Nterminal LBD | | Descriptor: | ARSENIC, Thyroid hormone receptor beta-1, [4-(4-HYDROXY-3-IODO-PHENOXY)-3,5-DIIODO-PHENYL]-ACETIC ACID | | Authors: | Huber, B.R, Desclozeaux, M, West, B.L, Cunha-Lima, S.T, Nguyen, H.T, Baxter, J.D, Ingraham, H.A, Fletterick, R.J. | | Deposit date: | 2003-01-20 | | Release date: | 2003-04-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Thyroid hormone receptor-beta mutations conferring hormone resistance and reduced corepressor release exhibit decreased stability in the N-terminal ligand-binding domain
Mol.Endocrinol., 17, 2003
|
|
2G5F
 
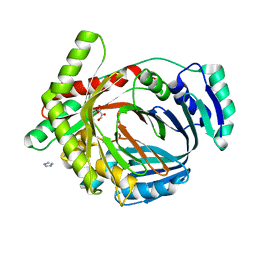 | | The structure of MbtI from Mycobacterium Tuberculosis, the first enzyme in the synthesis of Mycobactin, reveals it to be a salicylate synthase | | Descriptor: | COG0147: Anthranilate/para-aminobenzoate synthases component I, GLYCEROL, IMIDAZOLE, ... | | Authors: | Harrison, A.J, Lott, J.S, Yu, M, Ramsay, R, Baker, E.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-22 | | Release date: | 2006-09-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of MbtI from Mycobacterium tuberculosis, the First Enzyme in the Biosynthesis of the Siderophore Mycobactin, Reveals It To Be a Salicylate Synthase
J.Bacteriol., 188, 2006
|
|
4AAC
 
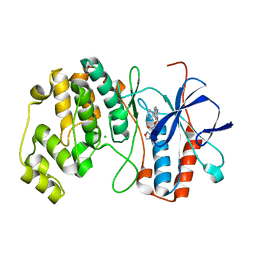 | | P38ALPHA MAP KINASE BOUND TO CMPD 29 | | Descriptor: | CHLORIDE ION, MITOGEN-ACTIVATED PROTEIN KINASE 14, N-isoxazol-3-yl-4-methyl-3-[6-(4-methylpiperazin-1-yl)-4-oxo-quinazolin-3-yl]benzamide | | Authors: | Gerhardt, S, Hargreaves, D. | | Deposit date: | 2011-12-01 | | Release date: | 2012-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of N-Cyclopropyl-4-Methyl-3-[6--(4-Methylpiperazin-1-Yl-4-Oxoquinazolin-3(4H)-Yl]Benzamide (Azd6703), a Clinical P38Alpha Map Kinase Inhibitor for the Treatment of Inflammatory Diseases
Bioorg.Med.Chem.Lett., 22, 2012
|
|
3MGY
 
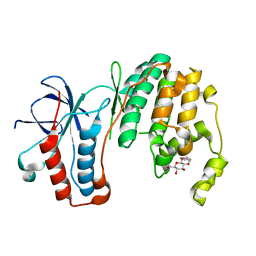 | | Mutagenesis of p38 MAP Kinase eshtablishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-out state | | Descriptor: | Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Namboodiri, H.V, Karpusas, M, Bukhtiyarova, M, Springman, E.B. | | Deposit date: | 2010-04-07 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mutagenesis of p38alpha MAP kinase establishes key roles of Phe169 in function and structural dynamics and reveals a novel DFG-OUT state.
Biochemistry, 46, 2007
|
|
2FOC
 
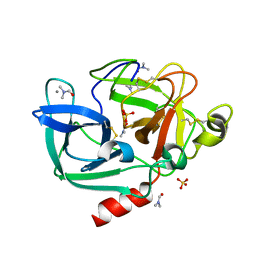 | | Structure of porcine pancreatic elastase in 55% dimethylformamide | | Descriptor: | CALCIUM ION, DIMETHYLFORMAMIDE, SULFATE ION, ... | | Authors: | Mattos, C, Bellamacina, C.R, Peisach, E, Pereira, A, Vitkup, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2006-01-13 | | Release date: | 2006-04-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Multiple solvent crystal structures: Probing binding sites, plasticity and hydration
J.Mol.Biol., 357, 2006
|
|
2JP1
 
 | |
2FYR
 
 | |
2JQI
 
 | |
7JV7
 
 | | Crystal Structure of the yeast RNA Pol II CTD kinase CTDK-1 complex | | Descriptor: | CITRATE ANION, CTD kinase subunit alpha, CTD kinase subunit beta, ... | | Authors: | Xie, Y, Ren, Y. | | Deposit date: | 2020-08-20 | | Release date: | 2021-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.850553 Å) | | Cite: | Structure and activation mechanism of the yeast RNA Pol II CTD kinase CTDK-1 complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7K7W
 
 | | The X-ray crystal structure of SSR4, an S. pombe chromatin remodelling protein: native | | Descriptor: | CHLORIDE ION, GLYCEROL, SWI/SNF and RSC complexes subunit ssr4 | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2020-09-24 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The X-ray crystal structure of the N-terminal domain of Ssr4, a Schizosaccharomyces pombe chromatin-remodelling protein.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2F4S
 
 | | A-site RNA in complex with neamine | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranoside, 5'-R(P*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
4PRG
 
 | | 0072 PARTIAL AGONIST PPAR GAMMA COCRYSTAL | | Descriptor: | (+/-)(2S,5S)-3-(4-(4-CARBOXYPHENYL)BUTYL)-2-HEPTYL-4-OXO-5-THIAZOLIDINE, PROTEIN (PEROXISOME PROLIFERATOR ACTIVATED RECEPTOR GAMMA) | | Authors: | Milburn, M.V. | | Deposit date: | 1999-05-07 | | Release date: | 1999-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A peroxisome proliferator-activated receptor gamma ligand inhibits adipocyte differentiation.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2JW6
 
 | | Solution structure of the DEAF1 MYND domain | | Descriptor: | Deformed epidermal autoregulatory factor 1 homolog, ZINC ION | | Authors: | Spadaccini, R, Perrin, H, Bottomley, M, Ansieu, S, Sattler, M. | | Deposit date: | 2007-10-08 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Retraction notice to "Structure and functional analysis of the MYND domain" [J. Mol. Biol. (2006) 358, 498-508].
J.Mol.Biol., 376, 2008
|
|
2K1O
 
 | |
4POJ
 
 | | Crystal structure of human Retinoid X Receptor alpha-ligand binding domain complex with 7-methyl UAB30 and the coactivator peptide GRIP-1 | | Descriptor: | (2E,4E,6Z,8E)-3,7-dimethyl-8-(7-methyl-3,4-dihydronaphthalen-1(2H)-ylidene)octa-2,4,6-trienoic acid, Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Xia, G, Smith, C.D, Muccio, D.D. | | Deposit date: | 2014-02-25 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Methyl substitution of a rexinoid agonist improves potency and reveals site of lipid toxicity.
J.Med.Chem., 57, 2014
|
|
1NLA
 
 | | Solution Structure of Switch Arc, a Mutant with 3(10) Helices Replacing a Wild-Type Beta-Ribbon | | Descriptor: | Transcriptional repressor arc | | Authors: | Cordes, M.H, Walsh, N.P, McKnight, C.J, Sauer, R.T. | | Deposit date: | 2003-01-06 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Switch Arc, a mutant with 3(10) helices replacing a wild-type beta-ribbon
J.Mol.Biol., 326, 2003
|
|
2F4T
 
 | | Asite RNA + designer antibiotic | | Descriptor: | (2R)-4-AMINO-N-{(1R,2S,3R,4R,5S)-5-AMINO-2-{2-[(2-AMINOETHYL)AMINO]ETHOXY}-4-[(2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-3-HYDROXYCYCLOHEXYL}-2-HYDROXYBUTANAMIDE, 5'-R(*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3', 5'-R(*UP*UP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*C)-3' | | Authors: | Murray, J.B, Meroueh, S.O, Russell, R.J, Lentzen, G, Haddad, J, Mobashery, S. | | Deposit date: | 2005-11-24 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interactions of designer antibiotics and the bacterial ribosomal aminoacyl-tRNA site
Chem.Biol., 13, 2006
|
|
