7BIJ
 
 | | Crystal Structure of SARS-CoV-2 main protease (Nsp5) in complex with compound 13 | | Descriptor: | (3~{S})-3'-(5-fluoranylpyridin-3-yl)spiro[1,2-dihydroindene-3,5'-imidazolidine]-2',4'-dione, 3C-like proteinase nsp5, DIMETHYL SULFOXIDE | | Authors: | Talibov, V.O. | | Deposit date: | 2021-01-12 | | Release date: | 2022-02-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses.
J.Am.Chem.Soc., 144, 2022
|
|
3C59
 
 | |
2GYU
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with HI-6 | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2GYW
 
 | | Crystal Structure of Mus musculus Acetylcholinesterase in Complex with Obidoxime | | Descriptor: | 1,1'-(OXYDIMETHYLENE)BIS(4-FORMYLPYRIDINIUM)DIOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2GJP
 
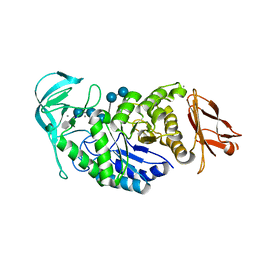 | | Structure of Bacillus halmapalus alpha-amylase, crystallized with the substrate analogue acarbose and maltose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lyhne-Iversen, L, Hobley, T.J, Kaasgaard, S.G, Harris, P. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Bacillus halmapalus alpha-amylase crystallized with and without the substrate analogue acarbose and maltose.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3BYD
 
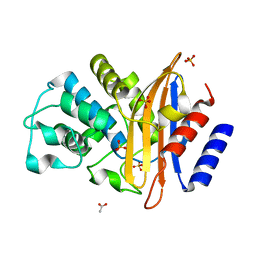 | | Crystal structure of beta-lactamase OXY-1-1 from Klebsiella oxytoca | | Descriptor: | ACETATE ION, Beta-lactamase OXY-1, SULFATE ION | | Authors: | Liang, Y.-H, Wu, S.W, Su, X.-D. | | Deposit date: | 2008-01-15 | | Release date: | 2009-01-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural insights into the broadened substrate profile of the extended-spectrum beta-lactamase OXY-1-1 from Klebsiella oxytoca
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2FF6
 
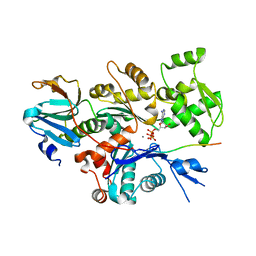 | | Crystal structure of Gelsolin domain 1:ciboulot domain 2 hybrid in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Aguda, A.H, Xue, B, Robinson, R.C. | | Deposit date: | 2005-12-19 | | Release date: | 2006-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structural Basis of Actin Interaction with Multiple WH2/beta-Thymosin Motif-Containing Proteins
Structure, 14, 2006
|
|
2GJR
 
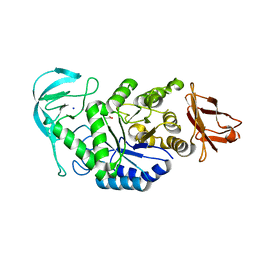 | | Structure of bacillus halmapalus alpha-amylase without any substrate analogues | | Descriptor: | ACETATE ION, CALCIUM ION, SODIUM ION, ... | | Authors: | Lyhne-Iversen, L, Hobley, T.J, Kaasgaard, S.G, Harris, P. | | Deposit date: | 2006-03-31 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Bacillus halmapalus alpha-amylase crystallized with and without the substrate analogue acarbose and maltose.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
3C9N
 
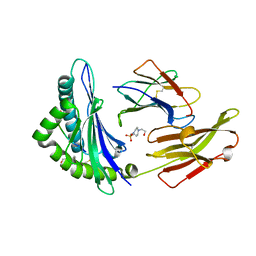 | | Crystal Structure of a SARS Corona Virus Derived Peptide Bound to the Human Major Histocompatibility Complex Class I molecule HLA-B*1501 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Roder, G.A, Kristensen, O, Kastrup, J.S, Buus, S, Gajhede, M. | | Deposit date: | 2008-02-18 | | Release date: | 2008-02-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
ACTA CRYSTALLOGR.,SECT.F, 64, 2008
|
|
3CBK
 
 | | chagasin-cathepsin B | | Descriptor: | Cathepsin B, Chagasin | | Authors: | Redzynia, I, Bujacz, G.D, Abrahamson, M, Ljunggren, A, Jaskolski, M, Mort, J.S. | | Deposit date: | 2008-02-22 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Displacement of the occluding loop by the parasite protein, chagasin, results in efficient inhibition of human cathepsin B.
J.Biol.Chem., 283, 2008
|
|
3CHR
 
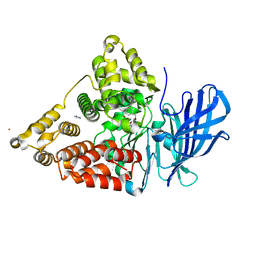 | | Crystal structure of leukotriene A4 hydrolase in complex with 4-amino-N-[4-(phenylmethoxy)phenyl]-butanamide | | Descriptor: | 4-amino-N-[4-(benzyloxy)phenyl]butanamide, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CZD
 
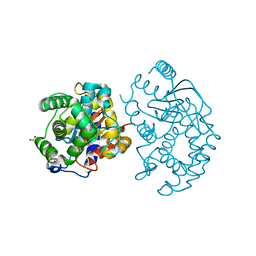 | | Crystal structure of human glutaminase in complex with L-glutamate | | Descriptor: | GLUTAMIC ACID, GLYCEROL, Glutaminase kidney isoform, ... | | Authors: | Karlberg, T, Welin, M, Andersson, J, Arrowsmith, C.H, Berglund, H, Busam, R.D, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wikstrom, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3CP7
 
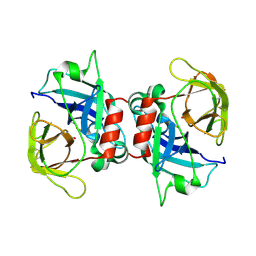 | |
3C5T
 
 | |
3CHS
 
 | | Crystal structure of leukotriene A4 hydrolase in complex with (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid | | Descriptor: | (2S)-2-amino-5-[[4-[(2S)-2-hydroxy-2-phenyl-ethoxy]phenyl]amino]-5-oxo-pentanoic acid, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
2GYV
 
 | | Crystal structure of Mus musculus Acetylcholinesterase in complex with Ortho-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15-PENTAOXAHEPTADECANE, ... | | Authors: | Pang, Y.P, Boman, M, Artursson, E, Akfur, C, Lundberg, S. | | Deposit date: | 2006-05-10 | | Release date: | 2006-08-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of acetylcholinesterase in complex with HI-6, Ortho-7 and obidoxime: Structural basis for differences in the ability to reactivate tabun conjugates.
Biochem.Pharm., 72, 2006
|
|
2EPO
 
 | | N-acetyl-B-D-glucosaminidase (GCNA) from Streptococcus gordonii | | Descriptor: | ACETIC ACID, N-acetyl-beta-D-glucosaminidase | | Authors: | Langley, D.B. | | Deposit date: | 2007-03-30 | | Release date: | 2008-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of N-acetyl-beta-D-glucosaminidase (GcnA) from the Endocarditis Pathogen Streptococcus gordonii and its Complex with the Mechanism-based Inhibitor NAG-thiazoline
J.Mol.Biol., 377, 2008
|
|
2HP3
 
 | | Crystal structure of iminodisuccinate epimerase | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IDS-epimerase, ... | | Authors: | Lohkamp, B, Bauerle, B, Rieger, P.G, Schneider, G. | | Deposit date: | 2006-07-17 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Three-dimensional Structure of Iminodisuccinate Epimerase Defines the Fold of the MmgE/PrpD Protein Family.
J.Mol.Biol., 362, 2006
|
|
2DY2
 
 | | Nitrite reductase pH 6.0 | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase | | Authors: | Jacobson, F. | | Deposit date: | 2006-09-05 | | Release date: | 2006-12-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | pH Dependence of Copper Geometry, Reduction Potential, and Nitrite Affinity in Nitrite Reductase
J.Biol.Chem., 282, 2007
|
|
2HK6
 
 | |
2HXB
 
 | |
2OYA
 
 | | Crystal structure analysis of the dimeric form of the SRCR domain of mouse MARCO | | Descriptor: | Macrophage receptor MARCO, SULFATE ION | | Authors: | Ojala, J.R.M, Pikkarainen, T, Tuuttila, A, Sandalova, T, Tryggvason, K. | | Deposit date: | 2007-02-21 | | Release date: | 2007-04-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the cysteine-rich domain of scavenger receptor MARCO reveals the presence of a basic and an acidic cluster that both contribute to ligand recognition.
J.Biol.Chem., 282, 2007
|
|
2OM1
 
 | |
2OO0
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, ACETATE ION, Ornithine decarboxylase, ... | | Authors: | Dufe, V.T, Ingner, D, Khomutov, A.R, Heby, O, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
4MI3
 
 | | Crystal structure of Gpb in complex with SUGAR (N-{(2R)-2-METHYL-3-[4-(PROPAN-2-YL)PHENYL]PROPANOYL}-BETA-D-GLUCOPYRANOSYLAMINE) (S21) | | Descriptor: | Glycogen phosphorylase, muscle form, N-{(2R)-2-methyl-3-[4-(propan-2-yl)phenyl]propanoyl}-beta-D-glucopyranosylamine | | Authors: | Kantsadi, L.A, Chatzileontiadou, S.M.D, Leonidas, D.D. | | Deposit date: | 2013-08-30 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure based inhibitor design targeting glycogen phosphorylase b. Virtual screening, synthesis, biochemical and biological assessment of novel N-acyl-beta-d-glucopyranosylamines.
Bioorg.Med.Chem., 22, 2014
|
|
