2KN5
 
 | |
4M5W
 
 | | Crystal structure of the USP7/HAUSP catalytic domain | | Descriptor: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Molland, K.L, Mesecar, A.D, Zhou, Q. | | Deposit date: | 2013-08-08 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4OUB
 
 | | A 2.20 angstroms X-ray crystal structure of E268A 2-aminomucaonate 6-semialdehyde dehydrogenase catalytic intermediate from Pseudomonas fluorescens | | Descriptor: | (2Z,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Huo, L, Liu, A. | | Deposit date: | 2014-02-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystallographic and spectroscopic snapshots reveal a dehydrogenase in action.
Nat Commun, 6, 2015
|
|
3AID
 
 | |
2JWO
 
 | | A PHD finger motif in the C-terminus of RAG2 modulates recombination activity | | Descriptor: | V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Ivanov, D, Hyberts, S.G, Sun, Z, Wagner, G. | | Deposit date: | 2007-10-17 | | Release date: | 2007-11-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A PHD finger motif in the C terminus of RAG2 modulates recombination activity.
J.Biol.Chem., 280, 2005
|
|
5T4I
 
 | |
5T5C
 
 | |
4KUK
 
 | | A superfast recovering full-length LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae (Dark state) | | Descriptor: | ACETIC ACID, RIBOFLAVIN, blue-light photoreceptor | | Authors: | Circolone, F, Granzin, J, Stadler, A, Krauss, U, Drepper, T, Endres, S, Knieps-Gruenhagen, E, Wirtz, A, Willbold, D, Batra-Safferling, R, Jaeger, K.-E. | | Deposit date: | 2013-05-22 | | Release date: | 2014-11-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of a short LOV protein from the marine phototrophic bacterium Dinoroseobacter shibae.
BMC Microbiol, 15, 2015
|
|
5T3V
 
 | |
1V1D
 
 | |
1CZ1
 
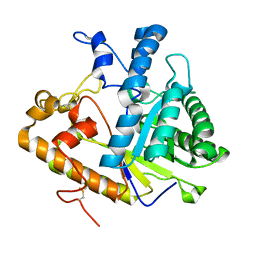 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS AT 1.85 A RESOLUTION | | Descriptor: | PROTEIN (EXO-B-(1,3)-GLUCANASE) | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 1999-09-01 | | Release date: | 2000-01-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
2G2I
 
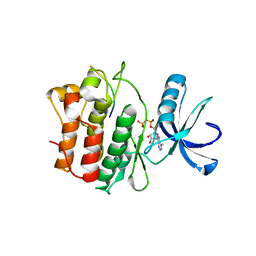 | |
3DGO
 
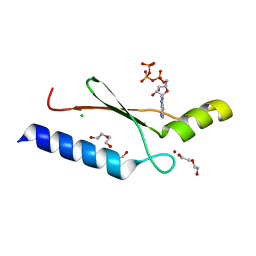 | | A non-biological ATP binding protein with a Tyr-Phe mutation in the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
1SZP
 
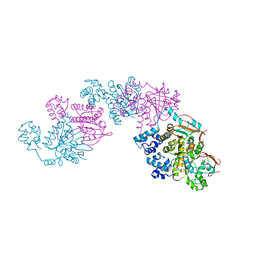 | | A Crystal Structure of the Rad51 Filament | | Descriptor: | DNA repair protein RAD51, SULFATE ION | | Authors: | Conway, A.B, Lynch, T.W, Zhang, Y, Fortin, G.S, Symington, L.S, Rice, P.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of a Rad51 filament.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1EZE
 
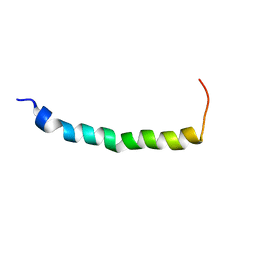 | | STRUCTURAL STUDIES OF A BABOON (PAPIO SP.) PLASMA PROTEIN INHIBITOR OF CHOLESTERYL ESTER TRANSFERASE. | | Descriptor: | CHOLESTERYL ESTER TRANSFERASE INHIBITOR PROTEIN | | Authors: | Buchko, G.W, Rozek, A, Kanda, P, Kennedy, M.A, Cushley, R.J. | | Deposit date: | 2000-05-10 | | Release date: | 2000-09-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of a baboon (Papio sp.) plasma protein inhibitor of cholesteryl ester transferase.
Protein Sci., 9, 2000
|
|
2DRE
 
 | | Crystal structure of Water-soluble chlorophyll protein from lepidium virginicum at 2.00 angstrom resolution | | Descriptor: | CHLOROPHYLL A, Water-soluble chlorophyll protein | | Authors: | Horigome, D, Satoh, H, Itoh, N, Mitsunaga, K, Oonishi, I, Nakagawa, A, Uchida, A. | | Deposit date: | 2006-06-08 | | Release date: | 2006-12-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural mechanism and photoprotective function of water-soluble chlorophyll-binding protein.
J.Biol.Chem., 282, 2007
|
|
2ZYS
 
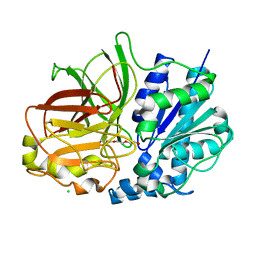 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2V3V
 
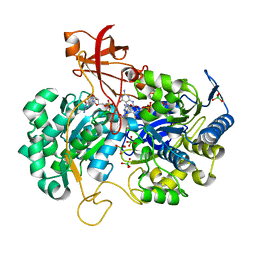 | | A New Catalytic Mechanism of Periplasmic Nitrate Reductase from Desulfovibrio desulfuricans ATCC 27774 from Crystallographic and EPR Data and based on detailed analysis of the sixth ligand | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, IRON/SULFUR CLUSTER, MOLYBDENUM ATOM, ... | | Authors: | Najmudin, S, Gonzalez, P.J, Trincao, J, Coelho, C, Mukhopadhyay, A, Romao, C.C, Moura, I, Moura, J.J, Brondino, C.D, Romao, M.J. | | Deposit date: | 2007-06-22 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Periplasmic Nitrate Reductase Revisited: A Sulfur Atom Completes the Sixth Coordination of the Catalytic Molybdenum.
J.Biol.Inorg.Chem., 13, 2008
|
|
3W21
 
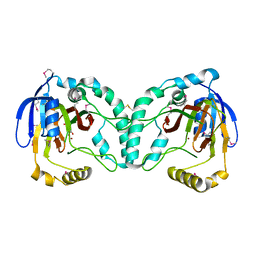 | | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase in complex with alpha-KG from Burkholderia ambifaria AMMD | | Descriptor: | 2-OXOGLUTARIC ACID, Putative uncharacterized protein, ZINC ION | | Authors: | Qin, H.M, Miyakawa, T, Jia, M.Z, Nakamura, A, Ohtsuka, J, Xue, Y.L, Kawashima, T, Kasahara, T, Hibi, M, Ogawa, J, Tanokura, M. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of a Novel N-Substituted L-Amino Acid Dioxygenase from Burkholderia ambifaria AMMD
Plos One, 8, 2013
|
|
3EY0
 
 | | A new form of DNA-drug interaction in the minor groove of a coiled coil | | Descriptor: | 1,5-BIS(4-AMIDINOPHENOXY)PENTANE, 5'-D(*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DTP*DAP*DT)-3', MAGNESIUM ION | | Authors: | Pous, J, Moreno, T, Subirana, J.A, Campos, J.L. | | Deposit date: | 2008-10-17 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Coiled-coil conformation of a pentamidine-DNA complex
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2W6C
 
 | | ACHE IN COMPLEX WITH A BIS-(-)-NOR-MEPTAZINOL DERIVATIVE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3R)-3-ethyl-1-{9-[(3S)-3-ethyl-3-(3-hydroxyphenyl)azepan-1-yl]nonyl}azepan-3-yl]phenol, ... | | Authors: | Paz, A, Xie, Q, Greenblatt, H.M, Fu, W, Tang, Y, Silman, I, Qiu, Z, Sussman, J.L. | | Deposit date: | 2008-12-18 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The Crystal Structure of a Complex of Acetylcholinesterase with a Bis-(-)-Nor-Meptazinol Derivative Reveals Disruption of the Catalytic Triad.
J.Med.Chem., 52, 2009
|
|
2BEQ
 
 | | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein | | Descriptor: | Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-11-29 | | Release date: | 2004-12-22 | | Last modified: | 2020-04-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2BEZ
 
 | | Structure of a proteolitically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein | | Descriptor: | GLYCEROL, Spike glycoprotein | | Authors: | Supekar, V.M, Bruckmann, C, Ingallinella, P, Bianchi, E, Pessi, A, Carfi, A. | | Deposit date: | 2004-12-02 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of a Proteolytically Resistant Core from the Severe Acute Respiratory Syndrome Coronavirus S2 Fusion Protein
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2ZYI
 
 | | A. Fulgidus lipase with fatty acid fragment and calcium | | Descriptor: | CALCIUM ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
1PXX
 
 | | CRYSTAL STRUCTURE OF DICLOFENAC BOUND TO THE CYCLOOXYGENASE ACTIVE SITE OF COX-2 | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kiefer, J.R, Rowlinson, S.W, Prusakiewicz, J.J, Pawlitz, J.L, Kozak, K.R, Kalgutkar, A.S, Stallings, W.C, Marnett, L.J, Kurumbail, R.G. | | Deposit date: | 2003-07-07 | | Release date: | 2003-09-09 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Novel Mechanism of Cyclooxygenase-2 Inhibition Involving Interactions with Ser-530 and Tyr-385.
J.Biol.Chem., 278, 2003
|
|
