6AS4
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 anti-crispr protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
5UZ9
 
 | | Cryo EM structure of anti-CRISPRs, AcrF1 and AcrF2, bound to type I-F crRNA-guided CRISPR surveillance complex | | Descriptor: | Anti-CRISPR protein 30, Anti-CRISPR protein Acr30-35, CRISPR RNA (60-MER), ... | | Authors: | Chowdhury, S, Carter, J, Rollins, M.F, Jackson, R.N, Hoffmann, C, Nosaka, L, Bondy-Denomy, J, Maxwell, K.L, Davidson, A.R, Fischer, E.R, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure Reveals Mechanisms of Viral Suppressors that Intercept a CRISPR RNA-Guided Surveillance Complex.
Cell, 169, 2017
|
|
4ZA0
 
 | | Structure of Human Enolase 2 in complex with Phosphonoacetohydroxamate | | Descriptor: | Gamma-enolase, MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID | | Authors: | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | Deposit date: | 2015-04-13 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
7M2X
 
 | | Open conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M3P
 
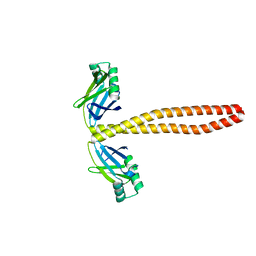 | | Xrcc4-Spc110p(164-207) fusion | | Descriptor: | Xrcc4-Spc110p(164-207) | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-18 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.0000186 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Y
 
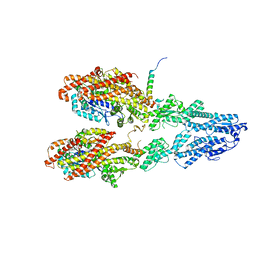 | | Closed conformation of the Yeast wild-type gamma-TuRC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2Z
 
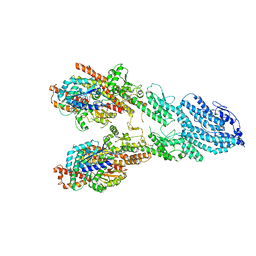 | | Monomeric single-particle reconstruction of the Yeast gamma-TuSC | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Spindle pole body component SPC97, Spindle pole body component SPC98, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
7M2W
 
 | | Engineered disulfide cross-linked closed conformation of the Yeast gamma-TuRC(SS) | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Spindle pole body component 110, Spindle pole body component SPC97, ... | | Authors: | Brilot, A.F, Lyon, A.S, Zelter, A, Viswanath, S, Maxwell, A, MacCoss, M.J, Muller, E.G, Sali, A, Davis, T.N, Agard, D.A. | | Deposit date: | 2021-03-17 | | Release date: | 2021-05-12 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | CM1-driven assembly and activation of yeast gamma-tubulin small complex underlies microtubule nucleation.
Elife, 10, 2021
|
|
4ZCW
 
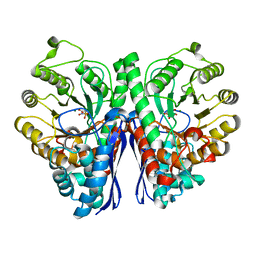 | | Structure of Human Enolase 2 in complex with SF2312 | | Descriptor: | Gamma-enolase, MAGNESIUM ION, [(3S,5S)-1,5-dihydroxy-2-oxopyrrolidin-3-yl]phosphonic acid | | Authors: | Leonard, P.G, Maxwell, D, Czako, B, Muller, F.L. | | Deposit date: | 2015-04-16 | | Release date: | 2016-07-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | SF2312 is a natural phosphonate inhibitor of enolase.
Nat.Chem.Biol., 12, 2016
|
|
7KIX
 
 | |
5BRL
 
 | | Crystal Structure of L124D STARD4 | | Descriptor: | StAR-related lipid transfer protein 4 | | Authors: | Iaea, D.B, Dikiy, I, Kiburu, I, Eliezer, D.E, Maxfield, F.R. | | Deposit date: | 2015-05-31 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | STARD4 Membrane Interactions and Sterol Binding.
Biochemistry, 54, 2015
|
|
7MSL
 
 | |
1ZS4
 
 | | Structure of bacteriophage lambda cII protein in complex with DNA | | Descriptor: | DNA - 27mer, Regulatory protein CII | | Authors: | Jain, D, Kim, Y, Maxwell, K.L, Beasley, S, Gussin, G.N, Edwards, A.M, Darst, S.A. | | Deposit date: | 2005-05-23 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Bacteriophage lambdacII and Its DNA Complex.
Mol.Cell, 19, 2005
|
|
1ZPQ
 
 | | STRUCTURE OF BACTERIOPHAGE LAMBDA CII protein | | Descriptor: | Regulatory protein CII | | Authors: | Jain, D, Kim, Y, Maxwell, K.L, Beasley, S, Gussin, G.N, Edwards, A.M, Joachimiak, A, Darst, S.A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2005-05-17 | | Release date: | 2005-08-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Bacteriophage lambdacII and Its DNA Complex.
Mol.Cell, 19, 2005
|
|
5W9D
 
 | |
5W9C
 
 | |
7M1N
 
 | |
7M1O
 
 | |
5GIO
 
 | | Crystal structure of box C/D RNP with 12 nt guide regions and 13 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.604 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5GIN
 
 | | Crystal structure of box C/D RNP with 12 nt guide regions and 9 nt substrates | | Descriptor: | 50S ribosomal protein L7Ae, C/D RNA, C/D box methylation guide ribonucleoprotein complex aNOP56 subunit, ... | | Authors: | Yang, Z, Lin, J, Ye, K. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.308 Å) | | Cite: | Box C/D guide RNAs recognize a maximum of 10 nt of substrates
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5NJA
 
 | | E. coli Microcin-processing metalloprotease TldD/E with angiotensin analogue bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, HIS-PRO-PHE, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NH3
 
 | | CRYSTAL STRUCTURE OF THE Activin receptor type-2A LIGAND BINDING DOMAIN IN COMPLEX WITH BIMAGRUMAB FV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Activin receptor type-2A, ... | | Authors: | Scheufler, C. | | Deposit date: | 2017-03-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Blockade of activin type II receptors with a dual anti-ActRIIA/IIB antibody is critical to promote maximal skeletal muscle hypertrophy.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5NJF
 
 | | E. coli Microcin-processing metalloprotease TldD/E (TldD H262A mutant) with pentapeptide bound | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALA-ALA-ALA-ALA-ALA, ... | | Authors: | Ghilarov, D, Serebryakova, M, Stevenson, C.E.M, Hearnshaw, S.J, Volkov, D, Maxwell, A, Lawson, D.M, Severinov, K. | | Deposit date: | 2017-03-28 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The Origins of Specificity in the Microcin-Processing Protease TldD/E.
Structure, 25, 2017
|
|
5NGV
 
 | |
