3URA
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V/S61T | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3URN
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/K185R/I274N/A80V/S61T with cyclohexyl methylphosphonate inhibitor | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
6IR0
 
 | | Zinc finger domain of the human DTX protein | | Descriptor: | Probable E3 ubiquitin-protein ligase DTX2, ZINC ION | | Authors: | Miyamoto, K. | | Deposit date: | 2018-11-09 | | Release date: | 2019-11-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Zinc finger domain of the human DTX protein adopts a unique RING fold.
Protein Sci., 28, 2019
|
|
2VJ3
 
 | | Human Notch-1 EGFs 11-13 | | Descriptor: | CALCIUM ION, CHLORIDE ION, NEUROGENIC LOCUS NOTCH HOMOLOG PROTEIN 1, ... | | Authors: | Johnson, S, Cordle, J, Tay, J.Z, Roversi, P, Lea, S.M. | | Deposit date: | 2007-12-06 | | Release date: | 2008-07-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Conserved Face of the Jagged/Serrate Dsl Domain is Involved in Notch Trans-Activation and Cis-Inhibition.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3M8Z
 
 | | Phosphopentomutase from Bacillus cereus bound with ribose-5-phosphate | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-O-phosphono-alpha-D-ribofuranose, ACETATE ION, ... | | Authors: | Panosian, T.D, Nannemann, D.P, Watkins, G, Wadzinski, B, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2010-03-19 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacillus cereus Phosphopentomutase Is an Alkaline Phosphatase Family Member That Exhibits an Altered Entry Point into the Catalytic Cycle.
J.Biol.Chem., 286, 2011
|
|
1FRQ
 
 | | FERREDOXIN:NADP+ OXIDOREDUCTASE (FERREDOXIN REDUCTASE) MUTANT E312A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION, PROTEIN (FERREDOXIN:NADP+ OXIDOREDUCTASE), ... | | Authors: | Aliverti, A, Deng, Z, Ravasi, D, Piubelli, L, Karplus, P.A, Zanetti, G. | | Deposit date: | 1998-10-10 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing the function of the invariant glutamyl residue 312 in spinach ferredoxin-NADP+ reductase.
J.Biol.Chem., 273, 1998
|
|
4AQ5
 
 | |
6OJD
 
 | |
6D3E
 
 | | PPARg LBD in Complex with SR1988 | | Descriptor: | 1-[(2,4-difluorophenyl)methyl]-2,3-dimethyl-N-[(1R)-1-phenylpropyl]-1H-indole-5-carboxamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Frkic, R.L, Bruning, J.B. | | Deposit date: | 2018-04-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structural and Dynamic Elucidation of a Non-acid PPARgammaPartial Agonist: SR1988.
Nucl Receptor Res, 5, 2018
|
|
3DRO
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN grown in ammonium sulfate | | Descriptor: | 2F5 Fab heavy chain, 2F5 Fab light chain, ELLELDKWASLWN gp41 peptide | | Authors: | Julien, J.-P, Bryson, S, Pai, E.F. | | Deposit date: | 2008-07-11 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3NB7
 
 | |
4QF5
 
 | | Crystal structure I of MurF from Acinetobacter baumannii | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, UDP-N-acetylmuramoyl-tripeptide--D-alanyl-D-alanine ligase | | Authors: | An, Y.J, Jeong, C.S, Cha, S.S. | | Deposit date: | 2014-05-19 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | ATP-binding mode including a carbamoylated lysine and two Mg(2+) ions, and substrate-binding mode in Acinetobacter baumannii MurF
Biochem.Biophys.Res.Commun., 450, 2014
|
|
3UR5
 
 | | Crystal Structure of PTE mutant K185R/I274N | | Descriptor: | COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, Parathion hydrolase | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
3URQ
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H with cyclohexyl methylphosphonate inhibitor | | Descriptor: | COBALT (II) ION, IMIDAZOLE, Parathion hydrolase, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-22 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
1QXE
 
 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | 5-HYDROXYMETHYL-FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
2ADM
 
 | | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI | | Descriptor: | ADENINE-N6-DNA-METHYLTRANSFERASE TAQI, S-ADENOSYLMETHIONINE | | Authors: | Schluckebier, G, Saenger, W. | | Deposit date: | 1996-07-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differential binding of S-adenosylmethionine S-adenosylhomocysteine and Sinefungin to the adenine-specific DNA methyltransferase M.TaqI.
J.Mol.Biol., 265, 1997
|
|
2H4W
 
 | |
3P0Q
 
 | | Human Tankyrase 2 - Catalytic PARP domain in complex with an inhibitor | | Descriptor: | N-[2-(4-chlorophenyl)ethyl]-6-methyl[1,2,4]triazolo[4,3-b]pyridazin-8-amine, SODIUM ION, SULFATE ION, ... | | Authors: | Karlberg, T, Siponen, M.I, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kotenyova, T, Kouznetsova, E, Moche, M, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Sehic, A, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Family-wide chemical profiling and structural analysis of PARP and tankyrase inhibitors
Nat.Biotechnol., 30, 2012
|
|
2VPM
 
 | | Trypanothione synthetase | | Descriptor: | BROMIDE ION, CHLORIDE ION, TRYPANOTHIONE SYNTHETASE | | Authors: | Fyfe, P.K, Oza, S.L, Fairlamb, A.H, Hunter, W.N. | | Deposit date: | 2008-03-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Leishmania Trypanothione Synthetase-Amidase Structure Reveals a Basis for Regulation of Conflicting Synthetic and Hydrolytic Activities.
J.Biol.Chem., 283, 2008
|
|
3URB
 
 | | Crystal Structure of PTE mutant H254G/H257W/L303T/M317L/I106C/F132I/L271I/K185R/I274N/A80V/R67H | | Descriptor: | COBALT (II) ION, DIETHYL HYDROGEN PHOSPHATE, IMIDAZOLE, ... | | Authors: | Tsai, P, Fox, N.G, Li, Y, Barondeau, D.P, Raushel, F.M. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymes for the homeland defense: optimizing phosphotriesterase for the hydrolysis of organophosphate nerve agents.
Biochemistry, 51, 2012
|
|
6FNJ
 
 | | Crystal Structure of Ephrin B4 (EphB4) Receptor Protein Kinase with an isomer of NVP-BHG712 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[(2-methyl-6-pyridin-3-yl-pyrazolo[3,4-d]pyrimidin-4-yl)amino]-~{N}-[3-(trifluoromethyl)phenyl]benzamide, Ephrin type-B receptor 4 | | Authors: | Kudlinzki, D, Troester, A, Witt, K, Linhard, V.L, Saxena, K, Schwalbe, H. | | Deposit date: | 2018-02-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.239 Å) | | Cite: | NVP-BHG712: Effects of Regioisomers on the Affinity and Selectivity toward the EPHrin Family.
ChemMedChem, 13, 2018
|
|
4AQ9
 
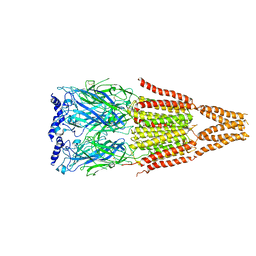 | |
1V2I
 
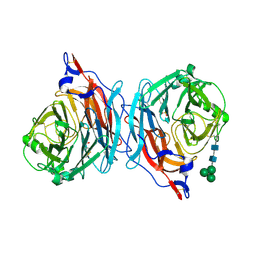 | | Structure of the hemagglutinin-neuraminidase from human parainfluenza virus type III | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Lawrence, M.C, Borg, N.A, Streltsov, V.A, Pilling, P.A, Epa, V.C, Varghese, J.N, McKimm-Breschkin, J.L, Colman, P.M. | | Deposit date: | 2003-10-16 | | Release date: | 2004-02-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Haemagglutinin-neuraminidase from Human Parainfluenza Virus Type III
J.Mol.Biol., 335, 2004
|
|
3K23
 
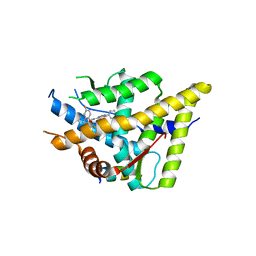 | | Glucocorticoid Receptor with Bound D-prolinamide 11 | | Descriptor: | 1-{[3-(4-{[(2R)-4-(5-fluoro-2-methoxyphenyl)-2-hydroxy-4-methyl-2-(trifluoromethyl)pentyl]amino}-6-methyl-1H-indazol-1-yl)phenyl]carbonyl}-D-prolinamide, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Biggadike, K.B, McLay, I.M, Madauss, K.P, Williams, S.P, Bledsoe, R.K. | | Deposit date: | 2009-09-29 | | Release date: | 2009-10-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Design and x-ray crystal structures of high-potency nonsteroidal glucocorticoid agonists exploiting a novel binding site on the receptor.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2C84
 
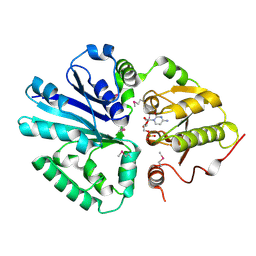 | | CRYSTAL STRUCTURE OF THE SIALYLTRANSFERASE PM0188 WITH CMP | | Descriptor: | ALPHA-2,3/2,6-SIALYLTRANSFERASE/SIALIDASE, CYTIDINE-5'-MONOPHOSPHATE | | Authors: | Kim, D.U, Cho, H.S. | | Deposit date: | 2005-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2019-10-09 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural analysis of sialyltransferase PM0188 from Pasteurella multocida complexed with donor analogue and acceptor sugar.
Bmb Rep, 41, 2008
|
|
