8CMK
 
 | | Transportin-3 TNPO3 in complex with RSY region of CIRBP | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, Cold-inducible RNA-binding protein, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhou, Q, Sagmeister, T, Pavkov-Keller, T, Madl, T. | | Deposit date: | 2023-02-20 | | Release date: | 2024-03-06 | | Last modified: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.945 Å) | | Cite: | Structural basis of phosphorylation-independent nuclear import of CIRBP by TNPO3.
Nat Commun, 16, 2025
|
|
6OMW
 
 | |
6OPB
 
 | |
2IRZ
 
 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | 3-{5-[(1R)-1-AMINO-1-METHYL-2-PHENYLETHYL]-1,3,4-OXADIAZOL-2-YL}-N-[(1R)-1-(4-FLUOROPHENYL)ETHYL]-5-[METHYL(METHYLSULFONYL)AMINO]BENZAMIDE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
4B1I
 
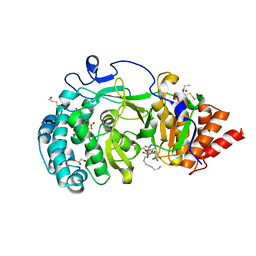 | | Structure of human PARG catalytic domain in complex with OA-ADP-HPD | | Descriptor: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 8-n-octylamino-adenosine diphosphate hydroxypyrrolidinediol, GLYCEROL, ... | | Authors: | Brassington, C, Ellston, J, Hassall, G, Holdgate, G, Johnson, T, McAlister, M, Smith, G, Tucker, J.A, Watson, M. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structures of the Human Poly (Adp-Ribose) Glycohydrolase Catalytic Domain Confirm Catalytic Mechanism and Explain Inhibition by Adp-Hpd Derivatives.
Plos One, 7, 2012
|
|
5TBP
 
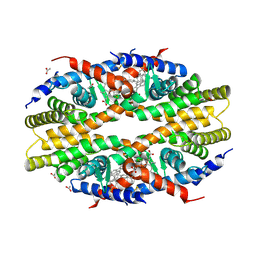 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
5US4
 
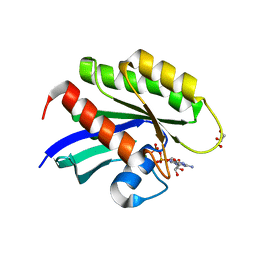 | | Crystal structure of human KRAS G12D mutant in complex with GDP | | Descriptor: | GLYCEROL, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T, Kaplan, A, Stockwell, B.R, Tong, L. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Multivalent Small-Molecule Pan-RAS Inhibitors.
Cell, 168, 2017
|
|
1I4Y
 
 | | THE CRYSTAL STRUCTURE OF PHASCOLOPSIS GOULDII WILD TYPE METHEMERYTHRIN | | Descriptor: | CHLORIDE ION, METHEMERYTHRIN, MU-OXO-DIIRON | | Authors: | Farmer, C.S, Kurtz Jr, D.M, Liu, Z.-J, Wang, B.C, Rose, J. | | Deposit date: | 2001-02-23 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of Phascolopsis gouldii wild type and L98Y methemerythrins: structural and functional alterations of the O2 binding pocket.
J.Biol.Inorg.Chem., 6, 2001
|
|
2IS0
 
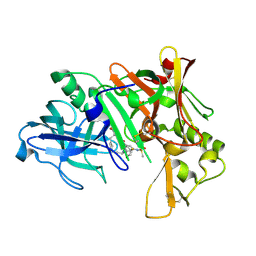 | | Crystal structure of human Beta-secretase complexed with inhibitor | | Descriptor: | (2S)-2-AMINO-2-BENZYL-3-HYDROXYPROPYL 3-({[(1R)-1-(4-FLUOROPHENYL)ETHYL]AMINO}CARBONYL)-5-[METHYL(METHYLSULFONYL)AMINO]BENZOATE, Beta-secretase 1 | | Authors: | Munshi, S. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of oxadiazoyl tertiary carbinamine inhibitors of beta-secretase (BACE-1).
J.Med.Chem., 49, 2006
|
|
9GFW
 
 | | HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH 4-methylphenyl-boronic acid at pH 7.4 | | Descriptor: | (4-methylphenyl)-tris(oxidanyl)boron, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2024-08-12 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Exploring the binding mode of phenyl and vinyl boronic acids to human carbonic anhydrases.
Int.J.Biol.Macromol., 282, 2024
|
|
1AZ0
 
 | | ECORV ENDONUCLEASE/DNA COMPLEX | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*TP*CP*TP*T)-3'), PROTEIN (TYPE II RESTRICTION ENZYME ECORV) | | Authors: | Perona, J.J, Martin, A.M. | | Deposit date: | 1997-11-24 | | Release date: | 1998-06-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational transitions and structural deformability of EcoRV endonuclease revealed by crystallographic analysis.
J.Mol.Biol., 273, 1997
|
|
6Q5G
 
 | |
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
4WW6
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2014-11-10 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
9GFX
 
 | | Huma Carbonic anhydrase II in complex with trans-2-phenylvinylboronic acid | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, ZINC ION, ... | | Authors: | Alterio, V, De Simone, G, Esposito, D. | | Deposit date: | 2024-08-12 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Exploring the binding mode of phenyl and vinyl boronic acids to human carbonic anhydrases.
Int.J.Biol.Macromol., 282, 2024
|
|
7TXS
 
 | | X-ray structure of the VioB N-aetyltransferase from Acinetobacter baumannii in the presence of a reaction intermediate | | Descriptor: | SODIUM ION, VioB, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)oxolan-2-yl]methyl (3R)-4-({3-[(2-{[(1S)-1-{[(2R,3S,4S,5R,6R)-4,5-dihydroxy-6-{[(R)-hydroxy{[(R)-hydroxy{[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)oxolan-2-yl]methoxy}phosphoryl]oxy}phosphoryl]oxy}-2-methyloxan-3-yl]amino}ethyl]sulfanyl}ethyl)amino]-3-oxopropyl}amino)-3-hydroxy-2,2-dimethyl-4-oxobutyl dihydrogen diphosphate (non-preferred name) | | Authors: | Herkert, N.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2022-02-09 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Structure and function of an N-acetyltransferase from the human pathogen Acinetobacter baumannii isolate BAL_212.
Proteins, 90, 2022
|
|
8QQM
 
 | | nicotinic acetylcholine receptor in intact synaptic membrane | | Descriptor: | Acetylcholine receptor subunit alpha, Acetylcholine receptor subunit beta, Acetylcholine receptor subunit delta, ... | | Authors: | Unwin, N. | | Deposit date: | 2023-10-05 | | Release date: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Influence of lipid bilayer on the structure of the muscle-type nicotinic acetylcholine receptor.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
9GFV
 
 | | HUMAN CARBONIC ANHYDRASE II IN COMPLEX WITH 4-methylphenyl-boronic acid | | Descriptor: | (4-methylphenyl)-tris(oxidanyl)boron, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Alterio, v, De Simone, G, Esposito, D. | | Deposit date: | 2024-08-12 | | Release date: | 2024-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Exploring the binding mode of phenyl and vinyl boronic acids to human carbonic anhydrases.
Int.J.Biol.Macromol., 282, 2024
|
|
6S83
 
 | | Crystal structure of methionine adenosyltransferase from Pyrococcus furiosus in complex with AMPPCP, SAM, and PCP | | Descriptor: | MAGNESIUM ION, METHYLENEDIPHOSPHONIC ACID, PHOSPHATE ION, ... | | Authors: | Degano, M, Minici, C, Porcelli, M. | | Deposit date: | 2019-07-08 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.336 Å) | | Cite: | Structures of catalytic cycle intermediates of the Pyrococcus furiosus methionine adenosyltransferase demonstrate negative cooperativity in the archaeal orthologues.
J.Struct.Biol., 210, 2020
|
|
7QYR
 
 | | Crystal structure of RimK from Pseudomonas aeruginosa PAO1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Probable alpha-L-glutamate ligase, poly-glutamate | | Authors: | Thompson, C.M.A, Little, R.H, Stevenson, C.E.M, Lawson, D.M, Malone, J.G. | | Deposit date: | 2022-01-29 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural insights into the mechanism of adaptive ribosomal modification by Pseudomonas RimK.
Proteins, 91, 2023
|
|
7UA6
 
 | | Horse liver alcohol dehydrogenase with NAD and trifluoroethanol at 25K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-11 | | Release date: | 2022-03-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UEI
 
 | | Horse liver alcohol dehydrogenase with NAD and pentafluorobenzyl alcohol at 100 K, 1.2 A, crystal 16 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2,3,4,5,6-PENTAFLUOROBENZYL ALCOHOL, Alcohol dehydrogenase E chain, ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-21 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
7UC9
 
 | | Horse liver alcohol dehydrogenase with NAD and trifluoroethanol at 45 K | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Alcohol dehydrogenase E chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), ... | | Authors: | Plapp, B.V, Gakhar, L. | | Deposit date: | 2022-03-16 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Dependence of crystallographic atomic displacement factors on temperature (25-150 K) for complexes of horse liver alcohol dehydrogenases
Acta Crystallogr.,Sect.D, D78, 2022
|
|
2WDB
 
 | | A family 32 carbohydrate-binding module, from the Mu toxin produced by Clostridium perfringens, in complex with beta-D-glcNAc-beta(1,2) mannose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose, CALCIUM ION, HYALURONOGLUCOSAMINIDASE | | Authors: | Ficko-Blean, E, Boraston, A.B. | | Deposit date: | 2009-03-23 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | N-Acetylglucosamine Recognition by a Family 32 Carbohydrate-Binding Module from Clostridium Perfringens Nagh.
J.Mol.Biol., 390, 2009
|
|
