4KYZ
 
 | | Three-dimensional structure of triclinic form of de novo design insertion domain, Northeast Structural Genomics Consortium (NESG) Target OR327 | | Descriptor: | Designed protein OR327 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Gleixner, J, Baker, D, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.492 Å) | | Cite: | Precise assembly of complex beta sheet topologies from de novo designed building blocks.
Elife, 4, 2015
|
|
4L7Z
 
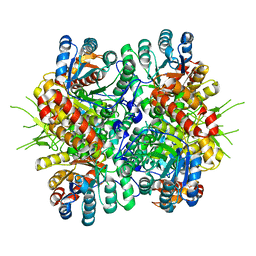 | | Crystal Structure of Chloroflexus aurantiacus malyl-CoA lyase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HpcH/HpaI aldolase | | Authors: | Zarzycki, J, Kerfeld, C.A. | | Deposit date: | 2013-06-14 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | The crystal structures of the tri-functional Chloroflexus aurantiacus and bi-functional Rhodobacter sphaeroides malyl-CoA lyases and comparison with CitE-like superfamily enzymes and malate synthases.
Bmc Struct.Biol., 13, 2013
|
|
4KZ9
 
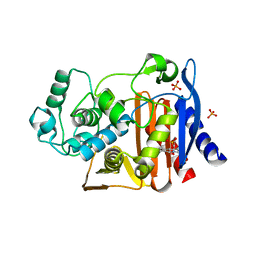 | | Crystal structure of AmpC beta-lactamase in complex with fragment 41 ((4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol) | | Descriptor: | (4R,4aS,8aS)-4-phenyldecahydroquinolin-4-ol, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Barelier, S, Fish, I, Shoichet, B.K. | | Deposit date: | 2013-05-29 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Increasing chemical space coverage by combining empirical and computational fragment screens.
Acs Chem.Biol., 9, 2014
|
|
4KZP
 
 | |
4L08
 
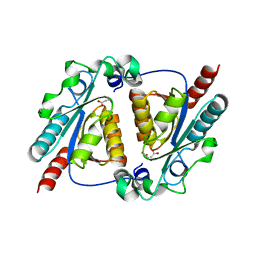 | | Crystal structure of the maleamate amidase Ami(C149A) in complex with maleate from Pseudomonas putida S16 | | Descriptor: | Hydrolase, isochorismatase family, MALEIC ACID | | Authors: | Chen, D.D, Lu, Y, Zhang, Z, Wu, G, Xu, P. | | Deposit date: | 2013-05-31 | | Release date: | 2014-07-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases.
Mol.Microbiol., 91, 2014
|
|
4KV0
 
 | |
4KVB
 
 | |
4KZ0
 
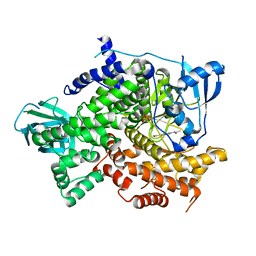 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION, methyl 2-(acetylamino)-1,3-benzothiazole-6-carboxylate | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZC
 
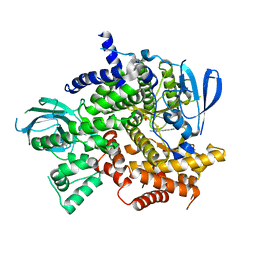 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4KZW
 
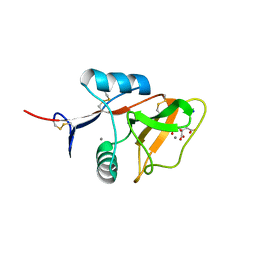 | | Structure of the carbohydrate-recognition domain of the C-type lectin mincle | | Descriptor: | C-TYPE LECTIN MINCLE, CALCIUM ION, CITRATE ANION, ... | | Authors: | Feinberg, H, Jegouzo, S.A.F, Rowntree, T.J.W, Guan, Y, Brash, M.A, Taylor, M.E, Weis, W.I, Drickamer, K. | | Deposit date: | 2013-05-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism for Recognition of an Unusual Mycobacterial Glycolipid by the Macrophage Receptor Mincle.
J.Biol.Chem., 288, 2013
|
|
4L06
 
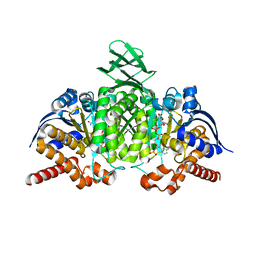 | |
4L1J
 
 | | Three dimensional structure of mutant D143A of human HD domain-containing protein 2, Northeast Structural Genomics Consortium (NESG) Target HR6723 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, HD domain-containing protein 2, ... | | Authors: | Kuzin, A, Su, M, Yakunin, A, Beloglazova, O, Seetharaman, J, Maglaqui, M, Xiao, R, Lee, D, Brown, G, Flick, R, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-03 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.824 Å) | | Cite: | Three dimensional structure of mutant D143A of human HD
domain-containing protein 2, Northeast Structural Genomics Consortium (NESG) Target HR6723
To be Published
|
|
4L1U
 
 | | Crystal Structure of Human Rtf1 Plus3 Domain in Complex with Spt5 CTR Phosphopeptide | | Descriptor: | GLYCEROL, RNA polymerase-associated protein RTF1 homolog, SULFATE ION, ... | | Authors: | Wier, A.D, Heroux, A, VanDemark, A.P. | | Deposit date: | 2013-06-03 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | Structural basis for Spt5-mediated recruitment of the Paf1 complex to chromatin.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4L46
 
 | | Crystal structures of human p70S6K1-WT | | Descriptor: | 2-{[4-(5-ethylpyrimidin-4-yl)piperazin-1-yl]methyl}-5-(trifluoromethyl)-1H-benzimidazole, RPS6KB1 protein, SULFATE ION, ... | | Authors: | Wang, J, Zhong, C, Ding, J. | | Deposit date: | 2013-06-07 | | Release date: | 2013-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structures of S6K1 provide insights into the regulation mechanism of S6K1 by the hydrophobic motif
Biochem.J., 454, 2013
|
|
4L5K
 
 | | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine | | Descriptor: | 6,7-dihydro[1,3]dioxolo[4,5-g][1,3]dioxolo[7,8]isoquino[3,2-a]isoquinolin-5-ium, DNA (5'-D(*CP*GP*AP*TP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P. | | Deposit date: | 2013-06-11 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex of DNA hexamer d(CGATCG) with Coptisine
to be published
|
|
4L7A
 
 | |
4L7I
 
 | | Crystal structure of S-Adenosylmethionine synthase from Sulfolobus solfataricus complexed with SAM and PPi | | Descriptor: | DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wang, F, Hurley, K.A, Helmich, K.E, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2013-06-13 | | Release date: | 2013-07-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Understanding molecular recognition of promiscuity of thermophilic methionine adenosyltransferase sMAT from Sulfolobus solfataricus.
Febs J., 281, 2014
|
|
4L83
 
 | |
4KUV
 
 | |
4L8C
 
 | | Crystal structure of the H2Db in complex with the NP-N3D peptide | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Rossjohn, J, Gras, S. | | Deposit date: | 2013-06-16 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Acute emergence and reversion of influenza A virus quasispecies within CD8(+) T cell antigenic peptides.
Nat Commun, 4, 2013
|
|
4KV9
 
 | | GTPase domain of Septin 10 from Schistosoma mansoni in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Septin | | Authors: | Zeraik, A.E, Pereira, H.M, Santos, Y.V, Brandao-Neto, J, Garratt, R.C, Araujo, A.P.U, Demarco, R. | | Deposit date: | 2013-05-22 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure of a Schistosoma mansoni Septin Reveals the Phenomenon of Strand Slippage in Septins Dependent on the Nature of the Bound Nucleotide.
J.Biol.Chem., 289, 2014
|
|
4L9V
 
 | |
4KXM
 
 | |
4KXV
 
 | | Human transketolase in covalent complex with donor ketose D-xylulose-5-phosphate, crystal 1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, D-XYLITOL-5-PHOSPHATE, ... | | Authors: | Neumann, P, Luedtke, S, Ficner, R, Tittmann, K. | | Deposit date: | 2013-05-28 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Sub-angstrom-resolution crystallography reveals physical distortions that enhance reactivity of a covalent enzymatic intermediate.
Nat Chem, 5, 2013
|
|
4LAG
 
 | | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines | | Descriptor: | 6-chloro-7-[5,6-dimethyl-2-(1,3-thiazol-2-yl)-1H-benzimidazol-1-yl]quinazoline-2,4-diamine, Dihydrofolate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hilgers, M.T. | | Deposit date: | 2013-06-19 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Design of New Dihydrofolate Reductase Antibacterial Agents: 7-(Benzimidazol-1-yl)-2,4-diaminoquinazolines.
J.Med.Chem., 57, 2014
|
|
