9N3N
 
 | |
9N3O
 
 | |
6UL8
 
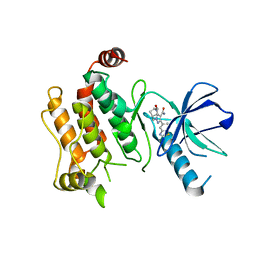 | | RIP2 kinase catalytic domain complex with (5S,6S,8R)-2-(benzo[d]thiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide | | Descriptor: | (5S,6S,8R)-2-(1,3-benzothiazol-5-yl)-6-hydroxy-4,5,6,7,8,9-hexahydro-5,8-methanopyrazolo[1,5-a][1,3]diazocine-3-carboxamide, CALCIUM ION, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Shewchuk, L.M, Convery, M.A. | | Deposit date: | 2019-10-07 | | Release date: | 2019-12-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Discovery of Pyrazolocarboxamides as Potent and Selective Receptor Interacting Protein 2 (RIP2) Kinase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6IAO
 
 | | Structure of Cytochrome P450 BM3 M11 mutant in complex with DTT at resolution 2.16A | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Bifunctional cytochrome P450/NADPH--P450 reductase, CHLORIDE ION, ... | | Authors: | Mirza, O, Rafiq, M, Frydenvang, K. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural analysis of Cytochrome P450 BM3 mutant M11 in complex with dithiothreitol.
Plos One, 14, 2019
|
|
8I5T
 
 | |
8I5U
 
 | |
7C6Z
 
 | | Crystal structure of beta-glycosides-binding protein (W67A) of ABC transporter in an open state | | Descriptor: | 1,2-ETHANEDIOL, CARBON DIOXIDE, CHLORIDE ION, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Samanta, R. | | Deposit date: | 2020-05-22 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Conformational Trapping of a beta-Glucosides-Binding Protein Unveils the Selective Two-Step Ligand-Binding Mechanism of ABC Importers.
J.Mol.Biol., 432, 2020
|
|
6BOZ
 
 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | Descriptor: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | Authors: | Babault, N, Anqi, M, Jin, J. | | Deposit date: | 2017-11-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
6ULA
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-cyclopropyl-N-{5-[4-({5-[(cyclopropylacetyl)amino]-1,3,4-thiadiazol-2-yl}oxy)piperidin-1-yl]-1,3,4-thiadiazol-2-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-07 | | Release date: | 2020-10-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00030
To Be Published
|
|
6UFO
 
 | |
2RFG
 
 | |
8RM5
 
 | | Cryo-EM structure of the cross-exon pre-B+5'ssLNG+ATPyS complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 5'SS oligo, NHP2-like protein 1, ... | | Authors: | Zhang, Z, Kumar, V, Dybkov, O, Will, C.L, Zhong, J, Ludwig, S, Urlaub, H, Kastner, B, Stark, H, Luehrmann, R. | | Deposit date: | 2024-01-05 | | Release date: | 2024-05-22 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Structural insights into the cross-exon to cross-intron spliceosome switch.
Nature, 630, 2024
|
|
2RJ0
 
 | |
8P2H
 
 | |
8B6R
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 labeled with a chloroalkane Cyanine3 fluorophore substrate | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Hellweg, L, Hiblot, J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A general method for the development of multicolor biosensors with large dynamic ranges.
Nat.Chem.Biol., 19, 2023
|
|
7CA4
 
 | |
5GWX
 
 | | Crystal structure of glycine sarcosine N-methyltransferase from Methanohalophilus portucalensis in complex with S-adenosylmethionine and sarcosine | | Descriptor: | 1,2-ETHANEDIOL, Glycine sarcosine N-methyltransferase, S-ADENOSYLMETHIONINE, ... | | Authors: | Lee, Y.R, Lin, T.S, Lai, S.J, Liu, M.S, Lai, M.C, Chan, N.L. | | Deposit date: | 2016-09-14 | | Release date: | 2016-11-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Structural Analysis of Glycine Sarcosine N-methyltransferase from Methanohalophilus portucalensis Reveals Mechanistic Insights into the Regulation of Methyltransferase Activity.
Sci Rep, 6, 2016
|
|
5ABH
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2R,3S,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
7KCP
 
 | |
6IAG
 
 | |
8P2G
 
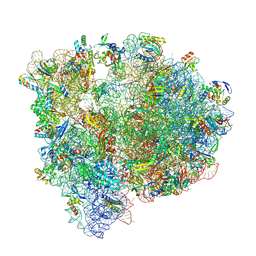 | | Staphylococcus aureus 70S ribosome with elongation factor G locked with fusidic acid cyclopentane with a tRNA in pe/E chimeric state | | Descriptor: | 1,4-DIAMINOBUTANE, 16S ribosomal RNA, 2-[(3~{R},4~{S},5~{S},8~{S},9~{S},10~{S},11~{R},13~{S},14~{S},16~{S})-16-acetyloxy-4,8,10,14-tetramethyl-3,11-bis(oxidanyl)-1,2,3,4,5,6,7,9,11,12,13,15,16,17-tetradecahydrocyclopenta[a]phenanthren-17-yl]-5-cyclopentyl-pentanoic acid, ... | | Authors: | Gonzalez-Lopez, A, Selmer, M. | | Deposit date: | 2023-05-16 | | Release date: | 2024-05-01 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structures of the Staphylococcus aureus ribosome inhibited by fusidic acid and fusidic acid cyclopentane.
Sci Rep, 14, 2024
|
|
8PEE
 
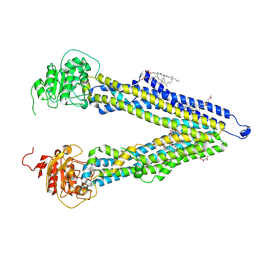 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
8P5C
 
 | | Crystal structure of the main protease (3CLpro/Mpro) of SARS-CoV-2 obtained in presence of 5 millimolar X77 enantiomer S. | | Descriptor: | 1,2-ETHANEDIOL, 3C-like proteinase nsp5, ACETATE ION, ... | | Authors: | Costanzi, E, Demitri, N, Storici, P. | | Deposit date: | 2023-05-23 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Unexpected Single-Ligand Occupancy and Negative Cooperativity in the SARS-CoV-2 Main Protease.
J.Chem.Inf.Model., 64, 2024
|
|
6D5M
 
 | | Ras:SOS:Ras in complex with a small molecule activator | | Descriptor: | 1-[(3-chloro-4-fluorophenyl)methyl]-5,6-dimethyl-2-(piperazin-1-yl)-1H-benzimidazole, GTPase HRas, MAGNESIUM ION, ... | | Authors: | Phan, J, Hodges, T, Fesik, S.W. | | Deposit date: | 2018-04-19 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Discovery and Structure-Based Optimization of Benzimidazole-Derived Activators of SOS1-Mediated Nucleotide Exchange on RAS.
J. Med. Chem., 61, 2018
|
|
3JTI
 
 | | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution | | Descriptor: | CALCIUM ION, Phospholipase A2 isoform 3, octapeptide from Amyloid beta A4 protein | | Authors: | Pandey, N, Mirza, Z, Vikram, G, Singh, N, Bhushan, A, Kaur, P, Srinivasan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2009-09-12 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex formed between Phospholipase A2 with beta-amyloid fragment, Lys-Gly-Ala-Ile-Ile-Gly-Leu-Met at 1.8 A resolution
To be Published
|
|
