5OSN
 
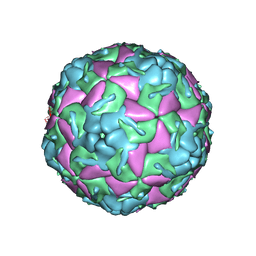 | | Crystal Structure of Bovine Enterovirus 2 determined with Serial Femtosecond X-ray Crystallography | | Descriptor: | Capsid protein, GLUTAMIC ACID, POTASSIUM ION, ... | | Authors: | Roedig, P, Ginn, H.M, Pakendorf, T, Sutton, G, Harlos, K, Walter, T.S, Meyer, J, Fischer, P, Duman, R, Vartiainen, I, Reime, B, Warmer, M, Brewster, A.S, Young, I.D, Michels-Clark, T, Sauter, N.K, Kotecha, A, Kelly, J, Rowlands, D.J, Sikorsky, M, Nelson, S, Damiani, D.S, Alonso-Mori, R, Ren, J, Fry, E.E, David, C, Stuart, D.I, Wagner, A, Meents, A. | | Deposit date: | 2017-08-17 | | Release date: | 2017-08-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-speed fixed-target serial virus crystallography.
Nat. Methods, 14, 2017
|
|
4Y30
 
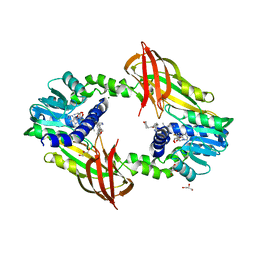 | | Crystal structure of human protein arginine methyltransferase PRMT6 bound to SAH and EPZ020411 | | Descriptor: | GLYCEROL, MAGNESIUM ION, N,N'-dimethyl-N-({3-[4-({trans-3-[2-(tetrahydro-2H-pyran-4-yl)ethoxy]cyclobutyl}oxy)phenyl]-1H-pyrazol-4-yl}methyl)ethane-1,2-diamine, ... | | Authors: | Swinger, K.K, Boriack-Sjodin, P.A. | | Deposit date: | 2015-02-10 | | Release date: | 2015-04-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Aryl Pyrazoles as Potent Inhibitors of Arginine Methyltransferases: Identification of the First PRMT6 Tool Compound.
Acs Med.Chem.Lett., 6, 2015
|
|
2RI3
 
 | |
2RJF
 
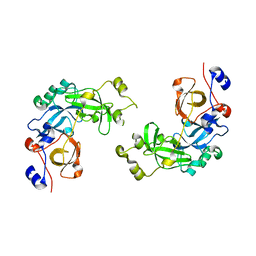 | | Crystal structure of L3MBTL1 in complex with H4K20Me2 (residues 12-30), orthorhombic form I | | Descriptor: | Histone H4, Lethal(3)malignant brain tumor-like protein | | Authors: | Allali-Hassani, A, Liu, Y, Herzanych, N, Ouyang, H, Mackenzie, F, Crombet, L, Loppnau, P, Kozieradzki, I, Vedadi, M, Weigelt, J, Sundstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J.R, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-14 | | Release date: | 2007-10-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | L3MBTL1 recognition of mono- and dimethylated histones.
Nat.Struct.Mol.Biol., 14, 2007
|
|
6E1Y
 
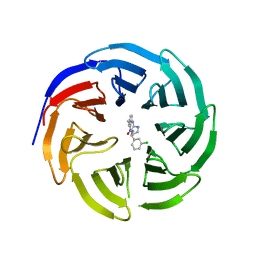 | |
6R6G
 
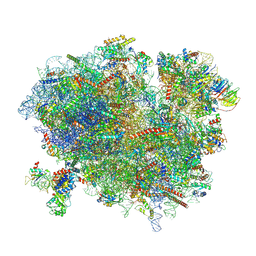 | | Structure of XBP1u-paused ribosome nascent chain complex with SRP. | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Braunger, K, Berninghausen, O, Beatrix, B, Beckmann, R. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
1U36
 
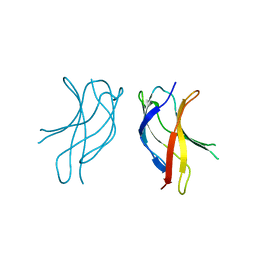 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
2RHZ
 
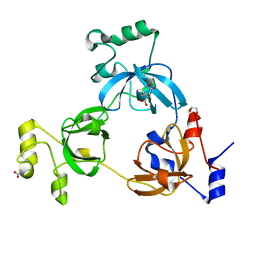 | |
6E1Z
 
 | |
7QPD
 
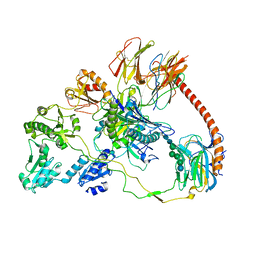 | | Structure of the human MHC I peptide-loading complex editing module | | Descriptor: | Beta-2-microglobulin, Calreticulin, HLA class I histocompatibility antigen, ... | | Authors: | Domnick, A, Susac, L, Trowitzsch, S, Thomas, C, Tampe, R. | | Deposit date: | 2022-01-03 | | Release date: | 2022-07-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Molecular basis of MHC I quality control in the peptide loading complex.
Nat Commun, 13, 2022
|
|
6E23
 
 | |
6E22
 
 | |
1U41
 
 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
5LSG
 
 | | PPARgamma complex with the betulinic acid | | Descriptor: | Betulinic Acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Pochetti, G, Montanari, R, Capelli, D, Loiodice, F, Laghezza, A, Calleri, E, Paiardini, A. | | Deposit date: | 2016-08-26 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Betulinic acid is a PPAR gamma antagonist that improves glucose uptake, promotes osteogenesis and inhibits adipogenesis.
Sci Rep, 7, 2017
|
|
7LMS
 
 | |
4Y2H
 
 | |
1U3Y
 
 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
6R5Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex (post-state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-07-10 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
8DG8
 
 | |
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
3W0A
 
 | | Crystal Structure Analysis of Vitamin D receptor | | Descriptor: | (4S)-4-hydroxy-5-[2-methyl-4-(3-{3-methyl-4-[4,4,4-trifluoro-3-hydroxy-3-(trifluoromethyl)but-1-yn-1-yl]phenyl}pentan-3-yl)phenoxy]pentanoic acid, Vitamin D3 receptor | | Authors: | Itoh, S, Iijima, S. | | Deposit date: | 2012-10-25 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | structure analysis of vitamin D3 receptor
To be Published
|
|
6TYS
 
 | | A potent cross-neutralizing antibody targeting the fusion glycoprotein inhibits Nipah virus and Hendra virus infection | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5B3 antibody heavy chain, ... | | Authors: | Dang, H.V, Chan, Y.P, Park, Y.J, Snijder, J, Da Silva, S.C, Vu, B, Yan, L, Feng, Y.R, Rockx, B, Geisbert, T, Mire, C.E, Broder, C.B, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-08-09 | | Release date: | 2019-10-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An antibody against the F glycoprotein inhibits Nipah and Hendra virus infections.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5LGA
 
 | | Structural analysis and biological activities of BXL0124, a Gemini analog of Vitamin D | | Descriptor: | (1~{R},3~{S},5~{Z})-5-[2-[(1~{R},3~{a}~{S},7~{a}~{R})-7~{a}-methyl-1-[(6~{R})-1,1,1-tris(fluoranyl)-10-methyl-2,10-bis(oxidanyl)-2-(trifluoromethyl)undeca-3,8-diyn-6-yl]-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, SRC-2, Vitamin D3 receptor A | | Authors: | Belorusova, A.Y, Rochel, N. | | Deposit date: | 2016-07-06 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and biological activities of BXL0124, a gemini analog of vitamin D.
J. Steroid Biochem. Mol. Biol., 173, 2017
|
|
4N23
 
 | | Crystal structure of the GP2 Core Domain from the California Academy of Science Virus, monoclinic symmetry | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GP2 Ectodomain | | Authors: | Malashkevich, V.N, Koellhoffer, J.F, Dai, Z, Toro, R, Lai, J.R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-04 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of the Glycoprotein GP2 Core Domain from the CAS Virus, a Novel Arenavirus-Like Species.
J.Mol.Biol., 426, 2014
|
|
