5VZQ
 
 | | Sperm whale myoglobin V68A/I107Y with nitric oxide | | Descriptor: | Myoglobin, NITRIC OXIDE, NITRITE ION, ... | | Authors: | Wang, B, Thomas, L.M, Richter-Addo, G.B. | | Deposit date: | 2017-05-29 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Nitrosyl Myoglobins and Their Nitrite Precursors: Crystal Structural and Quantum Mechanics and Molecular Mechanics Theoretical Investigations of Preferred Fe -NO Ligand Orientations in Myoglobin Distal Pockets.
Biochemistry, 57, 2018
|
|
4AOQ
 
 | | Cationic trypsin in complex with mutated Spinacia oleracea trypsin inhibitor III (SOTI-III) (F14A) | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, PENTAETHYLENE GLYCOL, ... | | Authors: | Schmelz, S, Glotzbach, B, Reinwarth, M, Christmann, A, Kolmar, H, Heinz, D.W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-01-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Spinacia Oleracea Trypsin Inhibitor III (Soti-III)
Acta Crystallogr.,Sect.D, 69, 2013
|
|
5VYT
 
 | | Crystal structure of the WbkC N-formyltransferase (F142A variant) from Brucella melitensis | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Gdp-mannose 4,6-dehydratase / gdp-4-amino-4,6-dideoxy-d-mannose formyltransferase, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
4PE5
 
 | | Crystal Structure of GluN1a/GluN2B NMDA Receptor Ion Channel | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(1R,2S)-2-(4-benzylpiperidin-1-yl)-1-hydroxypropyl]phenol, ... | | Authors: | Karakas, E, Furukawa, H. | | Deposit date: | 2014-04-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.96 Å) | | Cite: | Crystal structure of a heterotetrameric NMDA receptor ion channel.
Science, 344, 2014
|
|
4AH9
 
 | | Parallel screening of a low molecular weight compound library: do differences in methodology affect hit identification | | Descriptor: | 1,2-ETHANEDIOL, 1-(3-PHENYL-1,2,4-THIADIAZOL-5-YL)-1,4-DIAZEPANE, CHLORIDE ION, ... | | Authors: | Wielens, J, Heady, S.J, Rhodes, D.I, Mulder, R.J, Dolezal, O, Deadman, J.J, Newman, J, Chalmers, D.K, Parker, M.W, Peat, T.S, Scanlon, M.J. | | Deposit date: | 2012-02-06 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Screening of Low Molecular Weight Fragment Libraries: Do Differences in Methodology Affect Hit Identification?
J.Biomol.Screen, 18, 2013
|
|
7RZ3
 
 | |
5X7X
 
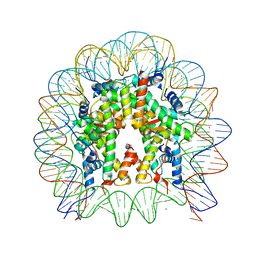 | | The crystal structure of the nucleosome containing H3.3 at 2.18 angstrom resolution | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-04-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.184 Å) | | Cite: | Crystal Structure and Characterization of Novel Human Histone H3 Variants, H3.6, H3.7, and H3.8
Biochemistry, 56, 2017
|
|
3AI4
 
 | | Crystal structure of yeast enhanced green fluorescent protein - mouse polymerase iota ubiquitin binding motif fusion protein | | Descriptor: | SULFATE ION, yeast enhanced green fluorescent protein,DNA polymerase iota | | Authors: | Suzuki, N, Wakatsuki, S, Kawasaki, M. | | Deposit date: | 2010-05-10 | | Release date: | 2010-09-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallization of small proteins assisted by green fluorescent protein
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5CZX
 
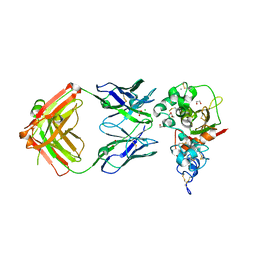 | | Crystal structure of Notch3 NRR in complex with 20358 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 20358 Fab heavy chain, ... | | Authors: | Hu, T, Fryer, C, Chopra, R, Clark, K. | | Deposit date: | 2015-08-01 | | Release date: | 2016-06-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of activating mutations of NOTCH3 in T-cell acute lymphoblastic leukemia and anti-leukemic activity of NOTCH3 inhibitory antibodies.
Oncogene, 35, 2016
|
|
4PSY
 
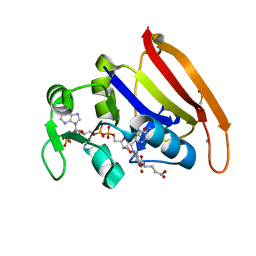 | | 100K crystal structure of Escherichia coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Wilson, M.A, Wan, Q, Bennet, B.C, Dealwis, C, Ringe, D, Petsko, G.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 22, 2014
|
|
5D0R
 
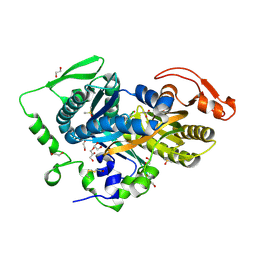 | | Crystal structure of human soluble Adenylyl Cyclase with the inhibitor bithionol | | Descriptor: | 1,2-ETHANEDIOL, 2,2'-sulfanediylbis(4,6-dichlorophenol), ACETATE ION, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2015-08-03 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Bithionol Potently Inhibits Human Soluble Adenylyl Cyclase through Binding to the Allosteric Activator Site.
J.Biol.Chem., 291, 2016
|
|
1K73
 
 | | Co-crystal Structure of Anisomycin Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, ANISOMYCIN, ... | | Authors: | Hansen, J, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-18 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
4PDJ
 
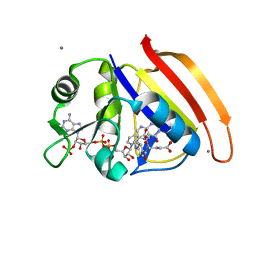 | | Neutron crystal Structure of E.coli Dihydrofolate Reductase complexed with folate and NADP+ | | Descriptor: | DIHYDROFOLIC ACID, Dihydrofolate reductase, MANGANESE (II) ION, ... | | Authors: | Wan, Q, Kovalevsky, A.Y, Wilson, M, Langan, P, Dealwis, C, Bennett, B. | | Deposit date: | 2014-04-18 | | Release date: | 2015-04-15 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.599 Å), X-RAY DIFFRACTION | | Cite: | Toward resolving the catalytic mechanism of dihydrofolate reductase using neutron and ultrahigh-resolution X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5XF0
 
 | |
5APN
 
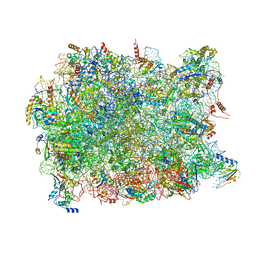 | | Structure of the yeast 60S ribosomal subunit in complex with Arx1, Alb1 and N-terminally tagged Rei1 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Greber, B.J, Gerhardy, S, Leitner, A, Leibundgut, M, Salem, M, Boehringer, D, Leulliot, N, Aebersold, R, Panse, V.G, Ban, V. | | Deposit date: | 2015-09-17 | | Release date: | 2015-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation.
Cell, 164, 2015
|
|
6V11
 
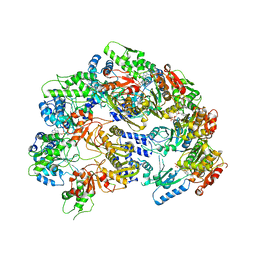 | | Lon Protease from Yersinia pestis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
1KNQ
 
 | | Crystal structure of gluconate kinase | | Descriptor: | CHLORIDE ION, Gluconate kinase | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-19 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
4R5Y
 
 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
1KNY
 
 | | KANAMYCIN NUCLEOTIDYLTRANSFERASE | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, KANAMYCIN A, KANAMYCIN NUCLEOTIDYLTRANSFERASE, ... | | Authors: | Pedersen, L.C, Benning, M.M, Holden, H.M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase.
Biochemistry, 34, 1995
|
|
5YTG
 
 | | Structure of large fragment of DNA Polymerase I from Thermus aquaticus Host-Guest complex with the unnatural base I-fC pair with dA | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(*AP*AP*AP*CP*GP*GP*CP*GP*CP*CP*GP*(94O)P*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Zeng, H, Mondal, M, Song, R.Y, Zhang, J, Xia, B, Gao, Y.Q, Yi, C.Q. | | Deposit date: | 2017-11-17 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Unnatural Cytosine Bases Recognized as Thymines by DNA Polymerases by the Formation of the Watson-Crick Geometry.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
5EMA
 
 | |
4RG6
 
 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
7S7P
 
 | |
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
5YJ4
 
 | |
