6XEO
 
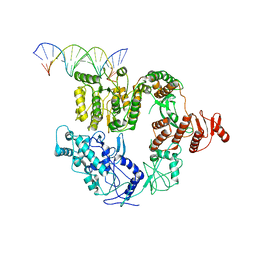 | | Structure of Mfd bound to dsDNA | | Descriptor: | DNA (5'-D(P*AP*GP*GP*AP*TP*AP*CP*TP*TP*AP*CP*AP*GP*CP*CP*AP*TP*C)-3'), DNA (5'-D(P*GP*AP*TP*GP*GP*CP*TP*GP*TP*AP*AP*GP*TP*AP*TP*CP*CP*T)-3'), Transcription-repair-coupling factor | | Authors: | Brugger, C, Deaconescu, A. | | Deposit date: | 2020-06-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Molecular determinants for dsDNA translocation by the transcription-repair coupling and evolvability factor Mfd.
Nat Commun, 11, 2020
|
|
6XKI
 
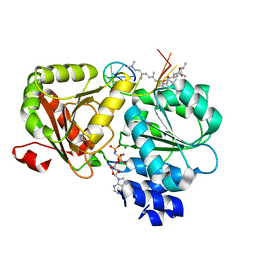 | | Crystal structure of eIF4A-I in complex with RNA bound to des-MePateA, a pateamine A analog | | Descriptor: | (3S,6Z,8E,11S,15R)-15-amino-3-[(1E,3E,5E)-7-(dimethylamino)-2,5-dimethylhepta-1,3,5-trien-1-yl]-9,11-dimethyl-4,12-dioxa-20-thia-21-azabicyclo[16.2.1]henicosa-1(21),6,8,18-tetraene-5,13-dione, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Liang, J, Naineni, S.K, Pelletier, J, Nagar, B. | | Deposit date: | 2020-06-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Functional mimicry revealed by the crystal structure of an eIF4A:RNA complex bound to the interfacial inhibitor, desmethyl pateamine A.
Cell Chem Biol, 28, 2021
|
|
7DD3
 
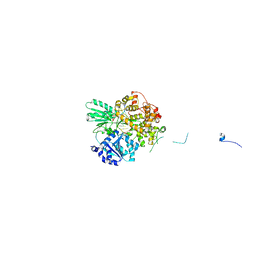 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCP
 
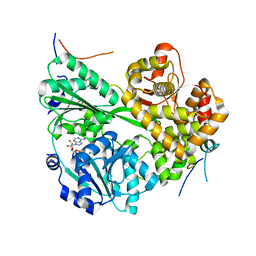 | | cryo-EM structure of the DEAH-box helicase Prp2 and coactivator Spp2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PRP2 isoform 1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7EVN
 
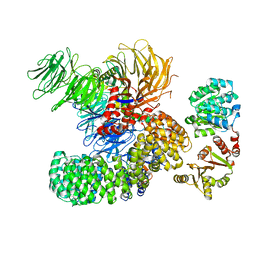 | | The cryo-EM structure of the DDX42-SF3b complex | | Descriptor: | ATP-dependent RNA helicase DDX42, PHD finger-like domain-containing protein 5A, Splicing factor 3B subunit 1, ... | | Authors: | Zhang, X, Zhan, X, Shi, Y. | | Deposit date: | 2021-05-21 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural insights into branch site proofreading by human spliceosome
Nat.Struct.Mol.Biol., 2024
|
|
8SCZ
 
 | |
8SD0
 
 | |
8SSW
 
 | |
8HUJ
 
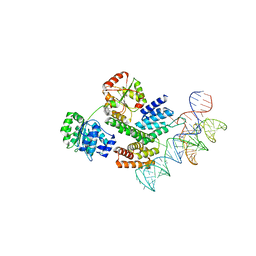 | | Cryo-EM structure of the J-K-St region of EMCV IRES in complex with eIF4G-HEAT1 and eIF4A | | Descriptor: | Eukaryotic initiation factor 4A-I, Eukaryotic translation initiation factor 4 gamma 1, IRES RNA (J-K-St), ... | | Authors: | Suzuki, H, Fujiyoshi, Y, Imai, S, Shimada, I. | | Deposit date: | 2022-12-24 | | Release date: | 2023-08-02 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Dynamically regulated two-site interaction of viral RNA to capture host translation initiation factor.
Nat Commun, 14, 2023
|
|
8T5S
 
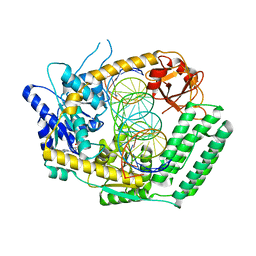 | | Cryo-EM structure of DRH-1 helicase and C-terminal domain bound to dsRNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dicer-related helicase, MAGNESIUM ION, ... | | Authors: | Consalvo, C.D, Donelick, H.M, Shen, P.S, Bass, B.L. | | Deposit date: | 2023-06-14 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Caenorhabditis elegans Dicer acts with the RIG-I-like helicase DRH-1 and RDE-4 to cleave dsRNA.
Elife, 13, 2024
|
|
8TBX
 
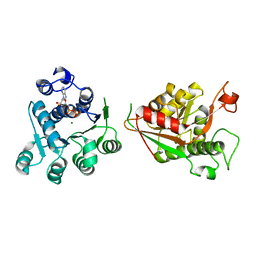 | | Crystal structure of human DDX1 helicase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase DDX1, MAGNESIUM ION, ... | | Authors: | Zeng, H, Dong, A, Li, Y, Yen, H, Hejazi, Z, Seitova, A, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2023-06-29 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of human DDX1 helicase in complex with ADP
To be published
|
|
8VXY
 
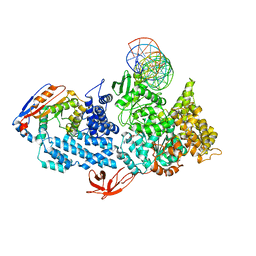 | | Structure of HamA(E138A,K140A)B-plasmid DNA complex from the Escherichia coli Hachiman defense system | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HamA, HamB, ... | | Authors: | Tuck, O.T, Hu, J.J, Doudna, J.A. | | Deposit date: | 2024-02-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Hachiman is a genome integrity sensor.
Biorxiv, 2024
|
|
8VXA
 
 | |
8VX9
 
 | |
8VXC
 
 | |
8V85
 
 | |
8W0A
 
 | |
5E7M
 
 | |
5DTU
 
 | |
7TO0
 
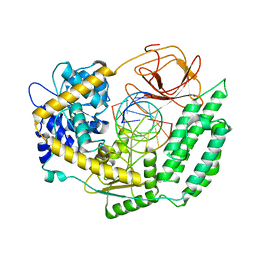 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
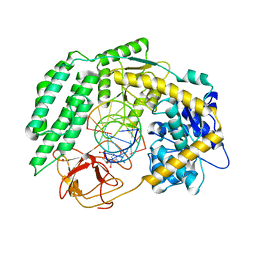 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | Descriptor: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-01-22 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TR8
 
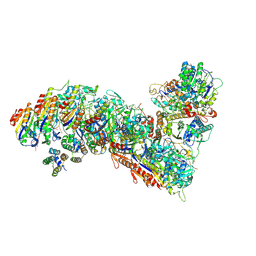 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
7TRA
 
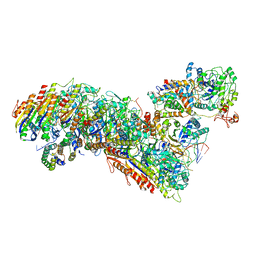 | | Cascade complex from type I-A CRISPR-Cas system | | Descriptor: | CRISPR-associated endonuclease Cas3-HD, CRISPR-associated helicase Cas3, Cas11a, ... | | Authors: | Hu, C, Ni, D, Nam, K.H, Majumdar, S, McLean, J, Stahlberg, H, Terns, M, Ke, A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Allosteric control of type I-A CRISPR-Cas3 complexes and establishment as effective nucleic acid detection and human genome editing tools.
Mol.Cell, 82, 2022
|
|
5F98
 
 | | Crystal structure of RIG-I in complex with Cap-0 RNA | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5F9F
 
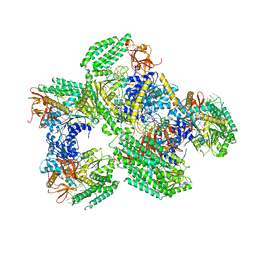 | | Crystal structure of RIG-I helicase-RD in complex with 24-mer blunt-end hairpin RNA | | Descriptor: | (R,R)-2,3-BUTANEDIOL, MAGNESIUM ION, Probable ATP-dependent RNA helicase DDX58, ... | | Authors: | Wang, C, Marcotrigiano, J, Miller, M.T, Jiang, F. | | Deposit date: | 2015-12-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for m7G recognition and 2'-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
