2XAS
 
 | | Crystal structure of LSD1-CoREST in complex with a tranylcypromine derivative (MC2580, 14e) | | Descriptor: | 3-[4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)PHENYL]PROPANOIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
6MZD
 
 | | Human TFIID Lobe A canonical | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (9.8 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6MZL
 
 | | Human TFIID canonical state | | Descriptor: | TATA-box-binding protein, Transcription initiation factor TFIID subunit 1, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
2GFA
 
 | | double tudor domain complex structure | | Descriptor: | Jumonji domain-containing protein 2A, peptide | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
5A1F
 
 | | Crystal structure of the catalytic domain of PLU1 in complex with N-oxalylglycine. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Srikannathasan, V, Johansson, C, Strain-Damerell, C, Gileadi, C, Szykowska, A, Kupinska, K, Kopec, J, Krojer, T, Steuber, H, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-04-29 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6OEO
 
 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5A3T
 
 | | Crystal structure of human PLU-1 (JARID1B) in complex with KDM5-C49 (2-(((2-((2-(dimethylamino)ethyl)(ethyl)amino)-2-oxoethyl)amino)methyl) isonicotinic acid). | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Kopec, J, Strain-Damerell, C, Kupinska, K, BurgessBrown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
5A1H
 
 | | Crystal structure of human Spindlin3 | | Descriptor: | SPINDLIN-3 | | Authors: | Srikannathasan, V, Gileadi, C, Johansson, C, Shrestha, L, Tallon, R, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-04-30 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Human Spindlin3
To be Published
|
|
5A3P
 
 | | Crystal structure of the catalytic domain of human PLU1 (JARID1B). | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, LYSINE-SPECIFIC DEMETHYLASE 5B, ... | | Authors: | Nowak, R, Srikannathasan, V, Johansson, C, Gileadi, C, Tallant, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, von Delft, F, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Oppermann, U. | | Deposit date: | 2015-06-02 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.008 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6OEQ
 
 | | Cryo-EM structure of mouse RAG1/2 12RSS-PRC/23RSS-NFC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OER
 
 | | Cryo-EM structure of mouse RAG1/2 NFC complex (DNA2) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2XAG
 
 | | Crystal structure of LSD1-CoREST in complex with para-bromo-(-)-trans- 2-phenylcyclopropyl-1-amine | | Descriptor: | 3-(4-BROMOPHENYL)PROPANAMIDE, FLAVIN-ADENINE DINUCLEOTIDE, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Binda, C, Valente, S, Romanenghi, M, Pilotto, S, Cirilli, R, Karytinos, A, Ciossani, G, Botrugno, O.A, Forneris, F, Tardugno, M, Edmondson, D.E, Minucci, S, Mattevi, A, Mai, A. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, Structural, and Biological Evaluation of Tranylcypromine Derivatives as Inhibitors of Histone Demethylases Lsd1 and Lsd2.
J.Am.Chem.Soc., 132, 2010
|
|
6WKR
 
 | | PRC2-AEBP2-JARID2 bound to H2AK119ub1 nucleosome | | Descriptor: | DNA (314-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Kasinath, V, Nogales, E, Beck, C, Sauer, P, Poepsel, S, Kosmatka, J, Faini, M, Toso, D, Aebersold, R. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-03 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | JARID2 and AEBP2 regulate PRC2 in the presence of H2AK119ub1 and other histone modifications.
Science, 371, 2021
|
|
2GF7
 
 | | Double tudor domain structure | | Descriptor: | Jumonji domain-containing protein 2A, SULFATE ION | | Authors: | Huang, Y, Fang, J, Bedford, M.T, Zhang, Y, Xu, R.M. | | Deposit date: | 2006-03-21 | | Release date: | 2006-05-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A
Science, 312, 2006
|
|
5CEH
 
 | | Structure of histone lysine demethylase KDM5A in complex with selective inhibitor | | Descriptor: | 7-oxo-5-phenyl-6-(propan-2-yl)-4,7-dihydropyrazolo[1,5-a]pyrimidine-3-carbonitrile, Lysine-specific demethylase 5A, NICKEL (II) ION, ... | | Authors: | Kiefer, J.R, Vinogradova, M. | | Deposit date: | 2015-07-06 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | An inhibitor of KDM5 demethylases reduces survival of drug-tolerant cancer cells.
Nat.Chem.Biol., 12, 2016
|
|
3O70
 
 | | PHD-type zinc finger of human PHD finger protein 13 | | Descriptor: | GLYCEROL, PHD finger protein 13, ZINC ION | | Authors: | Lam, R, Bian, C.B, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-29 | | Release date: | 2010-09-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.
Elife, 5, 2016
|
|
3QZT
 
 | | Crystal Structure of BPTF bromo in complex with histone H4K16ac - Form II | | Descriptor: | GLYCEROL, Histone H4, Nucleosome-remodeling factor subunit BPTF | | Authors: | Li, H, Ruthenburg, A.J, Patel, D.J. | | Deposit date: | 2011-03-07 | | Release date: | 2011-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Recognition of a Mononucleosomal Histone Modification Pattern by BPTF via Multivalent Interactions.
Cell(Cambridge,Mass.), 145, 2011
|
|
3QZS
 
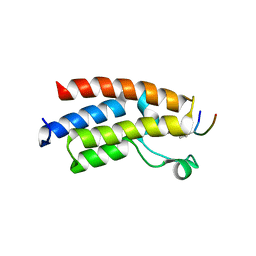 | |
3QLA
 
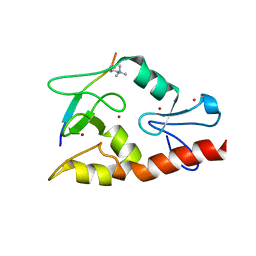 | | Hexagonal complex structure of ATRX ADD bound to H3K9me3 peptide | | Descriptor: | POTASSIUM ION, Transcriptional regulator ATRX, ZINC ION, ... | | Authors: | Xiang, B, Li, H. | | Deposit date: | 2011-02-02 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QLN
 
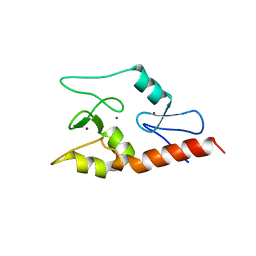 | | Crystal structure of ATRX ADD domain in free state | | Descriptor: | Transcriptional regulator ATRX, ZINC ION | | Authors: | Li, H, Patel, D.J. | | Deposit date: | 2011-02-03 | | Release date: | 2011-06-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QL9
 
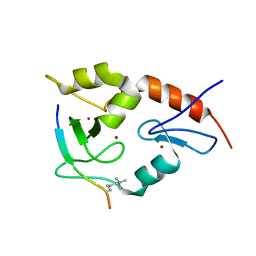 | |
3QLC
 
 | |
5KR7
 
 | | KDM4C bound to pyrazolo-pyrimidine scaffold | | Descriptor: | 6-ethyl-2,5-dimethyl-7-oxidanylidene-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bellon, S.F, Poy, F, Setser, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent, selective KDM5 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5L3C
 
 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5LUG
 
 | | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide | | Descriptor: | 1,2-ETHANEDIOL, ALA-ARG-THR-M3L-GLN-THR-ALA-2MR-LYS-SER, N3, ... | | Authors: | Srikannathasan, V, Gileadi, C, Talon, R, Shrestha, L, Kopec, J, Szykowska, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Oppermann, U, Huber, K. | | Deposit date: | 2016-09-08 | | Release date: | 2017-09-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Spindlin-2B protein in complex with ART(M3L)QTA(2MR)KS peptide
To be published
|
|
