4UFT
 
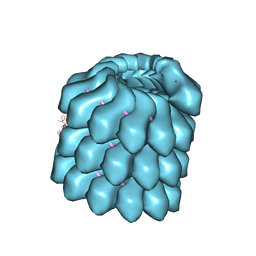 | | Structure of the helical Measles virus nucleocapsid | | Descriptor: | 5'-R(*CP*CP*CP*CP*CP*CP)-3', NUCLEOPROTEIN | | Authors: | Gutsche, I, Desfosses, A, Effantin, G, Ling, W.L, Haupt, M, Ruigrok, R.W.H, Sachse, C, Schoehn, G. | | Deposit date: | 2015-03-19 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Near-Atomic Cryo-Em Structure of the Helical Measles Virus Nucleocapsid.
Science, 348, 2015
|
|
4AIC
 
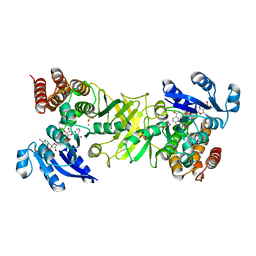 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin, manganese and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose- 5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JCV
 
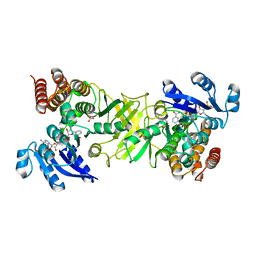 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
2JCX
 
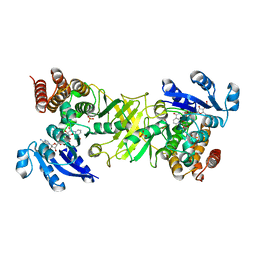 | | X-ray structure of mutant 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with fosmidomycin and NADPH | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Henriksson, L.M, Unge, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2007-01-04 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Mycobacterium Tuberculosis 1-Deoxy-D- Xylulose-5-Phosphate Reductoisomerase Provide New Insights Into Catalysis.
J.Biol.Chem., 282, 2007
|
|
9IL7
 
 | | Crystal Structure of SME E166A in Complex with Ceftaroline Fosamil | | Descriptor: | (2~{R})-2-[(1~{R})-1-[[(2~{Z})-2-ethoxyimino-2-[5-(phosphonoamino)-1,2,4-thiadiazol-3-yl]ethanoyl]amino]-2-oxidanylidene-ethyl]-5-[[4-(1-methylpyridin-4-yl)-1,3-thiazol-2-yl]sulfanyl]-3,6-dihydro-2~{H}-1,3-thiazine-4-carboxylic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Dhankhar, K, Hazra, S. | | Deposit date: | 2024-06-29 | | Release date: | 2025-07-02 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of SME E166A in Complex with Ceftaroline Fosamil
To Be Published
|
|
1JPY
 
 | | Crystal structure of IL-17F | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hymowitz, S.G, Filvaroff, E.H, Yin, J, Lee, J, Cai, L, Risser, P, Maruoka, M, Mao, W, Foster, J, Kelley, R, Pan, G, Gurney, A.L, de Vos, A.M, Starovasnik, M.A. | | Deposit date: | 2001-08-03 | | Release date: | 2001-09-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | IL-17s adopt a cystine knot fold: structure and activity of a novel cytokine, IL-17F, and implications for receptor binding.
EMBO J., 20, 2001
|
|
5I3U
 
 | |
3A06
 
 | | Crystal structure of DXR from Thermooga maritia, in complex with fosmidomycin and NADPH | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, ... | | Authors: | Takenoya, M, Ohtaki, A, Noguchi, K, Sasaki, Y, Ohsawa, K, Yohda, M, Yajima, S. | | Deposit date: | 2009-03-02 | | Release date: | 2010-03-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 1-deoxy-d-xylulose 5-phosphate reductoisomerase from the hyperthermophile Thermotoga maritima for insights into the coordination of conformational changes and an inhibitor binding
J.Struct.Biol., 170, 2010
|
|
3WQR
 
 | | Crystal structure of pfdxr complexed with inhibitor-12 | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CALCIUM ION, ... | | Authors: | Tanaka, N, Umeda, T. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC.
J.Med.Chem., 57, 2014
|
|
9BU5
 
 | | CC ProXp-ala apo solution structure | | Descriptor: | DNA-binding protein, putative | | Authors: | Duran, A.D, Foster, M.P. | | Deposit date: | 2024-05-16 | | Release date: | 2024-10-16 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | NMR-based solution structure of the Caulobacter crescentus ProXp-ala trans-editing enzyme.
Biomol.Nmr Assign., 18, 2024
|
|
9BE7
 
 | |
9BDS
 
 | |
9BE8
 
 | |
3WQQ
 
 | | Crystal structure of PfDXR complexed with inhibitor-3 | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, CALCIUM ION, ... | | Authors: | Tanaka, N, Umeda, T. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC.
J.Med.Chem., 57, 2014
|
|
5I42
 
 | | Structure of HIV-1 Reverse Transcriptase in complex with a DNA aptamer, AZTTP, and CA(2+) ion | | Descriptor: | 3'-AZIDO-3'-DEOXYTHYMIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (38-MER), ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2016-02-11 | | Release date: | 2016-06-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Conformational States of HIV-1 Reverse Transcriptase for Nucleotide Incorporation vs Pyrophosphorolysis-Binding of Foscarnet.
Acs Chem.Biol., 11, 2016
|
|
5ZDM
 
 | | The ligand-free structure of FomD | | Descriptor: | CALCIUM ION, FomD, GLYCEROL | | Authors: | Sato, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-02-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Biochemical and Structural Analysis of FomD That Catalyzes the Hydrolysis of Cytidylyl ( S)-2-Hydroxypropylphosphonate in Fosfomycin Biosynthesis.
Biochemistry, 57, 2018
|
|
5HRO
 
 | |
5ZDN
 
 | | The complex structure of FomD with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, FomD, GLYCEROL, ... | | Authors: | Sato, S, Miyanaga, A, Kudo, F, Eguchi, T. | | Deposit date: | 2018-02-23 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Biochemical and Structural Analysis of FomD That Catalyzes the Hydrolysis of Cytidylyl ( S)-2-Hydroxypropylphosphonate in Fosfomycin Biosynthesis.
Biochemistry, 57, 2018
|
|
3WQS
 
 | | Crystal structure of pfdxr complexed with inhibitor-126 | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, apicoplast, MAGNESIUM ION, ... | | Authors: | Tanaka, N, Umeda, T. | | Deposit date: | 2014-01-31 | | Release date: | 2014-11-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Binding Modes of Reverse Fosmidomycin Analogs toward the Antimalarial Target IspC.
J.Med.Chem., 57, 2014
|
|
7RHY
 
 | |
7RHZ
 
 | |
7RHX
 
 | |
8PHE
 
 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
8PHF
 
 | | Cryo-EM structure of human ACAD9-S191A | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
2Y1G
 
 | | X-ray structure of 1-deoxy-D-xylulose 5-phosphate reductoisomerase, DXR, Rv2870c, from Mycobacterium tuberculosis, in complex with a 3,4- dichlorophenyl-substituted FR900098 analogue and manganese. | | Descriptor: | 1-DEOXY-D-XYLULOSE 5-PHOSPHATE REDUCTOISOMERASE, 3-(N-HYDROXYACETAMIDO)-1-(3,4-DICHLOROPHENYL)PROPYLPHOSPHONIC ACID, MANGANESE (II) ION, ... | | Authors: | Henriksson, L.M, Larsson, A.M.S, Bergfors, T, Bjorkelid, C, Unge, T, Mowbray, S.L, Jones, T.A. | | Deposit date: | 2010-12-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Design, Synthesis and X-Ray Crystallographic Studies of Alpha-Aryl Substituted Fosmidomycin Analogues as Inhibitors of Mycobacterium Tuberculosis 1-Deoxy-D-Xylulose-5-Phosphate Reductoisomerase
J.Med.Chem, 54, 2011
|
|
