5J04
 
 | |
4MKS
 
 | | Crystal structure of enolase from Lactobacillus gasseri | | Descriptor: | CHLORIDE ION, Enolase 2, MAGNESIUM ION | | Authors: | Raghunathan, K, Harris, P.T, Spurbeck, R.R, Arvidson, C.G, Arvidson, D.N. | | Deposit date: | 2013-09-05 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Crystal structure of an efficacious gonococcal adherence inhibitor: An enolase from Lactobacillus gasseri.
Febs Lett., 588, 2014
|
|
2ONE
 
 | |
6L7D
 
 | | Mycobacterium tuberculosis enolase mutant - S42A | | Descriptor: | 1,2-ETHANEDIOL, 2-PHOSPHOGLYCERIC ACID, ACETATE ION, ... | | Authors: | Ahmad, M, Jha, B, Tiwari, S, Dwivedy, A, Biswal, B.K. | | Deposit date: | 2019-11-01 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural snapshots of Mycobacterium tuberculosis enolase reveal dual mode of 2PG binding and its implication in enzyme catalysis.
Iucrj, 10, 2023
|
|
8DG4
 
 | | Group A streptococcus Enolase K252A, K255A, K434A, K435A mutant | | Descriptor: | Enolase | | Authors: | Tjia-Fleck, S.C, Readnour, B.M, Castellino, F.J. | | Deposit date: | 2022-06-23 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | High-Resolution Single-Particle Cryo-EM Hydrated Structure of Streptococcus pyogenes Enolase Offers Insights into Its Function as a Plasminogen Receptor.
Biochemistry, 62, 2023
|
|
6J36
 
 | | crystal structure of Mycoplasma hyopneumoniae Enolase | | Descriptor: | Enolase, GLYCEROL, SULFATE ION | | Authors: | Chen, R, Zhang, S, Gan, R, Xie, X, Feng, Z, Wang, W, Ran, T, Zhang, W, Xiang, Q, Shao, G. | | Deposit date: | 2019-01-04 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Featured Species-Specific Loops Are Found in the Crystal Structure ofMhpEno, a Cell Surface Adhesin FromMycoplasma hyopneumoniae.
Front Cell Infect Microbiol, 9, 2019
|
|
5OHG
 
 | | enolase in complex with RNase E | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res., 46, 2018
|
|
1W6T
 
 | | Crystal Structure Of Octameric Enolase From Streptococcus pneumoniae | | Descriptor: | ENOLASE, MAGNESIUM ION, NONAETHYLENE GLYCOL | | Authors: | Ehinger, S, Schubert, W.-D, Bergmann, S, Hammerschmidt, S, Heinz, D.W. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmin(Ogen)-Binding Alpha-Enolase from Streptococcus Pneumoniae: Crystal Structure and Evaluation of Plasmin(Ogen)-Binding Sites
J.Mol.Biol., 343, 2004
|
|
4ROP
 
 | |
6O4N
 
 | |
1IYX
 
 | | Crystal structure of enolase from Enterococcus hirae | | Descriptor: | ENOLASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Hosaka, T, Meguro, T, Yamato, I, Shirakihara, Y. | | Deposit date: | 2002-09-12 | | Release date: | 2003-07-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Enterococcus hirae Enolase at 2.8 A Resolution
J.BIOCHEM.(TOKYO), 133, 2003
|
|
5IDZ
 
 | |
8UEL
 
 | | Crystal structure of enolase from Litopenaeus vannamei | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Chang, X, Zhao, G. | | Deposit date: | 2023-10-01 | | Release date: | 2023-12-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Characterization and Structural Analyses of Enolase from Shrimp ( Litopenaeus vannamei ).
J.Agric.Food Chem., 71, 2023
|
|
6BFZ
 
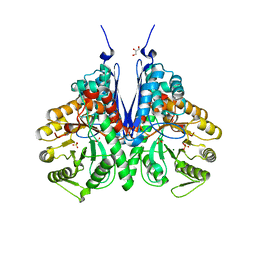 | | Crystal structure of enolase from E. coli with a mixture of apo form, substrate, and product form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-PHOSPHOGLYCERIC ACID, Enolase, ... | | Authors: | Erlandsen, H, Wright, D, Krucinska, J. | | Deposit date: | 2017-10-27 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and Functional Studies of Bacterial Enolase, a Potential Target against Gram-Negative Pathogens.
Biochemistry, 58, 2019
|
|
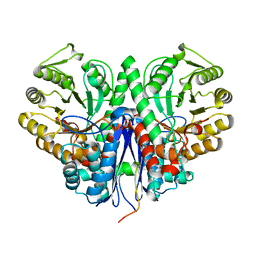
3H8A
 
 | |
5WRO
 
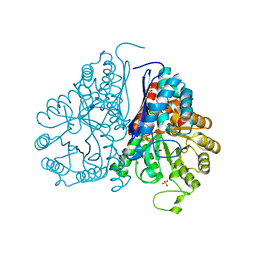 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
2FYM
 
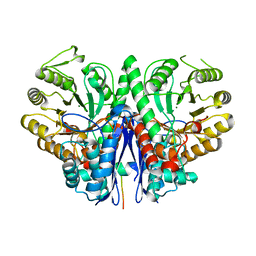 | |
7MBH
 
 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
6NPF
 
 | | Structure of E.coli enolase in complex with an analog of the natural product SF-2312 metabolite. | | Descriptor: | Enolase, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Erlandsen, H, Krucinska, J, Lombardo, M, Wright, D. | | Deposit date: | 2019-01-17 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Functional and structural basis of E. coli enolase inhibition by SF2312: a mimic of the carbanion intermediate.
Sci Rep, 9, 2019
|
|
6NB2
 
 | |
7N6G
 
 | | C1 of central pair | | Descriptor: | CPC1, Calmodulin, DPY30, ... | | Authors: | Han, L, Zhang, K. | | Deposit date: | 2021-06-08 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of an active central apparatus.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5ENL
 
 | |
5EU9
 
 | | Structure of Human Enolase 2 in complex with ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid | | Descriptor: | ((3S,5S)-1,5-dihydroxy-3-methyl-2-oxopyrrolidin-3-yl)phosphonic acid, Gamma-enolase, MAGNESIUM ION, ... | | Authors: | Leonard, P.G, Muller, F.L. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | SF2312, a natural phosphonate inhibitor of Enolase
To be Published
|
|
3B97
 
 | | Crystal Structure of human Enolase 1 | | Descriptor: | Alpha-enolase, MAGNESIUM ION, SULFATE ION | | Authors: | Kang, H.J, Jung, S.K, Kim, S.J, Chung, S.J. | | Deposit date: | 2007-11-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of human alpha-enolase (hENO1), a multifunctional glycolytic enzyme.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3ENL
 
 | |
