6D36
 
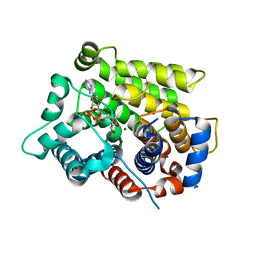 | | Structure of human ARH3 bound to ADP-ribose and magnesium | | Descriptor: | MAGNESIUM ION, Poly(ADP-ribose) glycohydrolase ARH3, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Pourfarjam, Y, Ventura, J, Kurinov, I, Kim, I.K. | | Deposit date: | 2018-04-14 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human ADP-ribosyl-acceptor hydrolase 3 bound to ADP-ribose reveals a conformational switch that enables specific substrate recognition.
J.Biol.Chem., 293, 2018
|
|
8SGB
 
 | |
6Q7W
 
 | |
1IFL
 
 | |
1J6O
 
 | |
6NZD
 
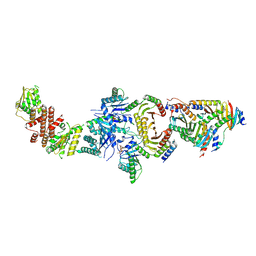 | | Cryo-EM Structure of the Lysosomal Folliculin Complex (FLCN-FNIP2-RagA-RagC-Ragulator) | | Descriptor: | 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(thiophosphonooxy)phosphoryl]oxy}phosphoryl]-alpha-L-arabinofuranosyl}-3,9-dihydro-1H-purine-2,6-dione, Folliculin, Folliculin-interacting protein 2, ... | | Authors: | Fromm, S.A, Young, L.N, Hurley, J.H. | | Deposit date: | 2019-02-13 | | Release date: | 2019-11-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex.
Science, 366, 2019
|
|
6Q7V
 
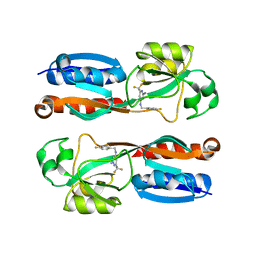 | |
2REW
 
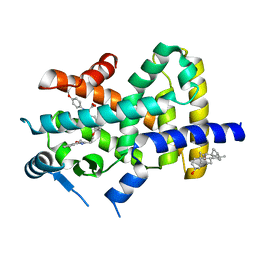 | | Crystal Structure of PPARalpha ligand binding domain with BMS-631707 | | Descriptor: | (2S,3S)-1-(4-METHOXYPHENYL)-3-(3-(2-(5-METHYL-2-PHENYLOXAZOL-4-YL)ETHOXY)BENZYL)-4-OXOAZETIDINE-2-CARBOXYLIC ACID, N,N-BIS(3-D-GLUCONAMIDOPROPYL)DEOXYCHOLAMIDE, Peroxisome proliferator-activated receptor alpha | | Authors: | Muckelbauer, J. | | Deposit date: | 2007-09-27 | | Release date: | 2007-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of Azetidinone Acids as Conformationally-Constrained Dual (alpha/gamma) PPAR Activators
To be Published
|
|
4G63
 
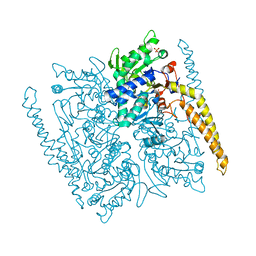 | | Crystal structure of cytosolic IMP-GMP specific 5'-nucleotidase (lpg0095) in complex with phosphate ions from Legionella pneumophila, Northeast Structural Genomics Consortium Target LgR1 | | Descriptor: | Cytosolic IMP-GMP specific 5'-nucleotidase, PHOSPHATE ION | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Ho, C.K, Ciccosanti, C, Mao, L, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-07-18 | | Release date: | 2012-10-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Allosteric regulation and substrate activation in cytosolic nucleotidase II from Legionella pneumophila.
Febs J., 281, 2014
|
|
6O22
 
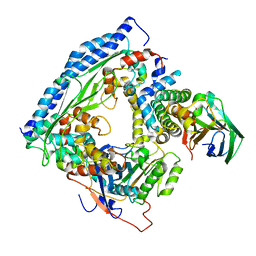 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
3R00
 
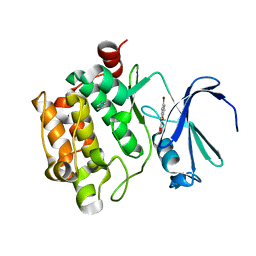 | |
3NRV
 
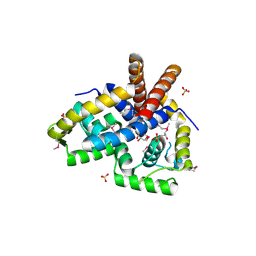 | |
3R02
 
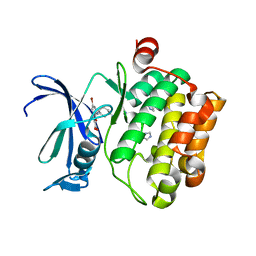 | | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors | | Descriptor: | 7-[(cis-4-aminocyclohexyl)amino]-5-bromo-1-benzofuran-2-carboxylic acid, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Xiang, Y, Hirth, B, Asmussen, G, Biemann, H.-P, Good, A, Fitzgerald, M, Gladysheva, T, Jancsics, K, Liu, J, Metz, M, Papoulis, A, Skerlj, R, Stepp, D.J, Wei, R.R. | | Deposit date: | 2011-03-07 | | Release date: | 2011-05-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The discovery of novel benzofuran-2-carboxylic acids as potent Pim-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3NRG
 
 | |
4HX7
 
 | |
3OLF
 
 | |
3ON2
 
 | | Structure of a protein with unknown function from Rhodococcus sp. RHA1 | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Probable transcriptional regulator, SULFATE ION | | Authors: | Fan, Y, Evdokimova, E, Egorova, O, Savchenko, A, Joachimiak, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-27 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of a protein with unknown function from Rhodococcus sp. RHA1
To be Published
|
|
3OOP
 
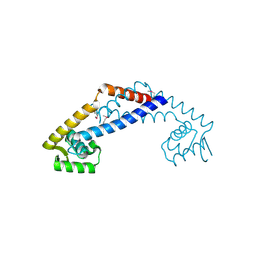 | | The structure of a protein with unknown function from Listeria innocua Clip11262 | | Descriptor: | Lin2960 protein | | Authors: | Fan, Y, Li, H, Zhou, Y, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a protein with unknown function from Listeria innocua Clip11262
To be Published
|
|
3OOF
 
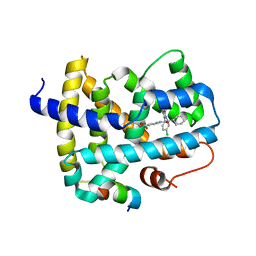 | |
2V7C
 
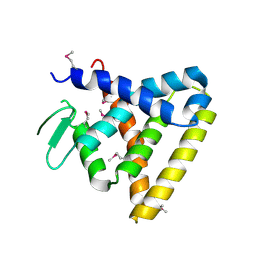 | | Crystal Structure of Rev-Erb beta | | Descriptor: | ORPHAN NUCLEAR RECEPTOR NR1D2 | | Authors: | Woo, E.-J, Jeong, D.G, Lim, M.-Y, Kim, S.J, Ryu, S.E. | | Deposit date: | 2007-07-29 | | Release date: | 2007-10-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Insight Into the Constitutive Repression Function of the Nuclear Receptor Rev-Erbbeta
J.Mol.Biol., 373, 2007
|
|
4HX8
 
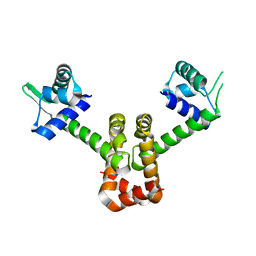 | | Structure of metal-free MNTR mutant E11K | | Descriptor: | Transcriptional regulator MntR | | Authors: | Glasfeld, A, Mcguire, A. | | Deposit date: | 2012-11-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Roles of the A and C Sites in the Manganese-Specific Activation of MntR.
Biochemistry, 52, 2013
|
|
4HV6
 
 | |
3T9I
 
 | | Pim1 complexed with a novel 3,6-disubstituted indole at 2.6 Ang Resolution | | Descriptor: | 2-methoxy-4-(3-phenyl-2H-pyrazolo[3,4-b]pyridin-6-yl)phenol, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Bellamacina, C, Shu, W, Le, V, Nishiguchi, G, Bussiere, D. | | Deposit date: | 2011-08-02 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of novel 3,5-disubstituted indole derivatives as potent inhibitors of Pim-1, Pim-2, and Pim-3 protein kinases.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3JYA
 
 | | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors | | Descriptor: | 6,9-dichloro[1]benzothieno[3,2-d]pyrimidin-4(3H)-one, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Stoll, V.S. | | Deposit date: | 2009-09-21 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of 3H-benzo[4,5]thieno[3,2-d]pyrimidin-4-ones as Potent, Highly Selective and Orally Bioavailable Pim Kinases Inhibitors
TO BE PUBLISHED
|
|
3JW4
 
 | | The structure of a putative MarR family transcriptional regulator from Clostridium acetobutylicum | | Descriptor: | GLYCEROL, IMIDAZOLE, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Bigelow, L, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a putative MarR family transcriptional regulator from Clostridium acetobutylicum
TO BE PUBLISHED
|
|
