1DI4
 
 | |
2P8L
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELLELDKWASLWN | | Descriptor: | gp41 peptide, nmAb 2F5, heavy chain, ... | | Authors: | Julien, J.P, Bryson, S, Pai, E.F. | | Deposit date: | 2007-03-22 | | Release date: | 2007-05-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
6VCU
 
 | | Homo sapiens FKBP12 protein bound with APX879 in P32 space group | | Descriptor: | ACETATE ION, N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
4YCU
 
 | | Crystal structure of cladosporin in complex with human lysyl-tRNA synthetase | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, GLYCEROL, LYSINE, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by an ATP Competitive Inhibitor.
Chem. Biol., 22, 2015
|
|
8FXE
 
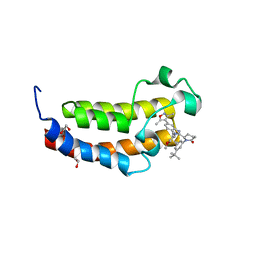 | | Bromodomain of CBP liganded with iCBP6 | | Descriptor: | (6S)-1-(3-tert-butylphenyl)-6-{(5P)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXA
 
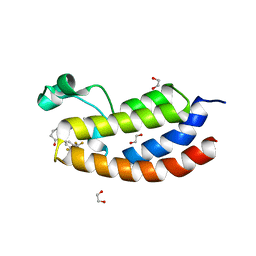 | | Bromodomain of CBP liganded with iCBP4 | | Descriptor: | (6S)-1-[3,5-bis(trifluoromethyl)phenyl]-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, 1,2-ETHANEDIOL, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-24 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
8FXN
 
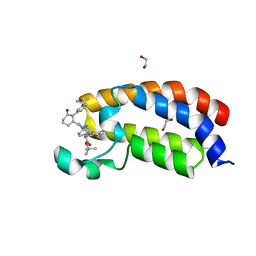 | | Bromodomain of CBP liganded with iCBP7 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, tert-butyl {(1R,4s)-4-[(5M)-2-[(2S)-1-(3-tert-butylphenyl)-6-oxopiperidin-2-yl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1H-benzimidazol-1-yl]cyclohexyl}carbamate | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks are sensitive to EP300/CBP bromodomain inhibition
To be published
|
|
8FXO
 
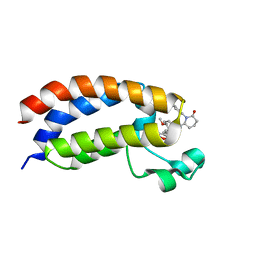 | | Bromodomain of CBP liganded with iCBP8 | | Descriptor: | (6S)-1-(2,3-dihydro-1,4-benzodioxin-6-yl)-6-{(5M)-5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[(1s,4R)-4-methoxycyclohexyl]-1H-benzimidazol-2-yl}piperidin-2-one, CREB-binding protein | | Authors: | Schonbrunn, E, Bikowitz, M. | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Group 3 medulloblastoma transcriptional networks collapse under domain specific EP300/CBP inhibition.
Nat Commun, 15, 2024
|
|
1DVN
 
 | |
4YCV
 
 | |
4YE6
 
 | |
4LHL
 
 | |
4LML
 
 | | GLIC double mutant I9'A T25'A | | Descriptor: | Proton-gated ion channel | | Authors: | Grosman, C, Gonzalez-Gutierrez, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1DF8
 
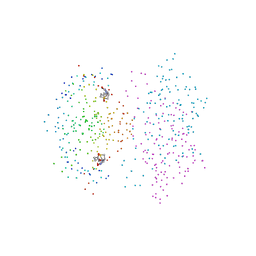 | | S45A MUTANT OF STREPTAVIDIN IN COMPLEX WITH BIOTIN | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Hyre, D.E, Le Trong, I, Freitag, S, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 1999-11-18 | | Release date: | 2000-09-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Ser45 plays an important role in managing both the equilibrium and transition state energetics of the streptavidin-biotin system.
Protein Sci., 9, 2000
|
|
5CLU
 
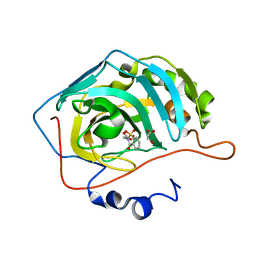 | | THE CRYSTAL STRUCTURE OF THE COMPLEX of HCAII WITH A SACCHARINE DERIVATIVE | | Descriptor: | (1,1-dioxido-3-oxo-1,2-benzothiazol-2(3H)-yl)acetic acid, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | D'Ambrosio, K, De Simone, G. | | Deposit date: | 2015-07-16 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A Combined Crystallographic and Theoretical Study Explains the Capability of Carboxylic Acids to Adopt Multiple Binding Modes in the Active Site of Carbonic Anhydrases.
Chemistry, 22, 2016
|
|
4PG3
 
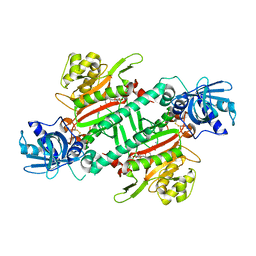 | | Crystal structure of KRS complexed with inhibitor | | Descriptor: | LYSINE, Lysine--tRNA ligase, cladosporin | | Authors: | Sharma, A, Yogavel, M, Khan, S, Sharma, A, Belrhali, H. | | Deposit date: | 2014-05-01 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.696 Å) | | Cite: | Structural basis of malaria parasite lysyl-tRNA synthetase inhibition by cladosporin.
J. Struct. Funct. Genomics, 15, 2014
|
|
6VCT
 
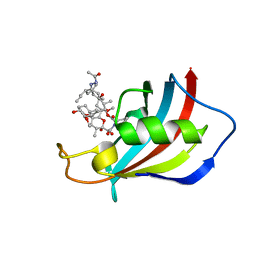 | | Mucor circinelloides FKBP12 protein bound with APX879 in C2221 space group | | Descriptor: | N'-[(3S,4R,5S,8R,9E,12S,14S,15R,16S,18R,19R,26aS)-5,19-dihydroxy-3-{(1E)-1-[(1R,3R,4R)-4-hydroxy-3-methoxycyclohexyl]prop-1-en-2-yl}-14,16-dimethoxy-4,10,12,18-tetramethyl-1,20,21-trioxo-8-(prop-2-en-1-yl)-1,3,4,5,6,8,11,12,13,14,15,16,17,18,19,20,21,23,24,25,26,26a-docosahydro-7H-15,19-epoxypyrido[2,1-c][1,4]oxazacyclotricosin-7-ylidene]acetohydrazide, Peptidylprolyl isomerase | | Authors: | Gobeil, S, Spicer, L. | | Deposit date: | 2019-12-23 | | Release date: | 2020-12-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Leveraging Fungal and Human Calcineurin-Inhibitor Structures, Biophysical Data, and Dynamics To Design Selective and Nonimmunosuppressive FK506 Analogs.
Mbio, 12, 2021
|
|
2EGO
 
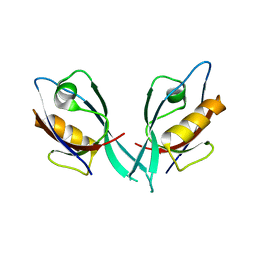 | | Crystal Structure of Tamalin PDZ Domain | | Descriptor: | General receptor for phosphoinositides 1-associated scaffold protein | | Authors: | Sugi, T, Oyama, T, Muto, T, Nakanishi, S, Morikawa, K, Jingami, H. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of autoinhibitory PDZ domain of Tamalin: implications for metabotropic glutamate receptor trafficking regulation
Embo J., 26, 2007
|
|
4YCW
 
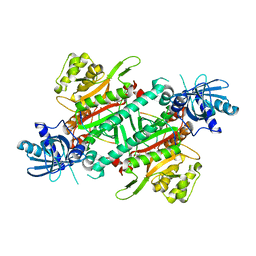 | | Crystal structure of cladosporin in complex with plasmodium like human lysyl-tRNA synthetase mutant | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 2, LYSINE, Lysine--tRNA ligase, ... | | Authors: | Fang, P, Wang, J, Guo, M. | | Deposit date: | 2015-02-20 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Basis for Specific Inhibition of tRNA Synthetase by ATP Competitive Inhibitor
Chem.Biol., 22, 2015
|
|
6M0T
 
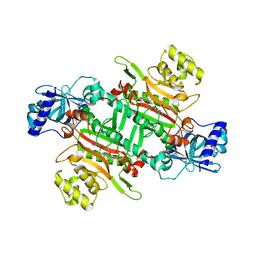 | | Crystal Structure of Lysyl-tRNA Synthetase from Plasmodium falciparum complexed with L-lysine and Cladosporin derivative (CL-2) | | Descriptor: | (3R)-3-[(R)-[(2R,6S)-6-methyloxan-2-yl]-oxidanyl-methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, LYSINE, Lysine--tRNA ligase | | Authors: | Babbar, P, Sharma, A, Manickam, Y. | | Deposit date: | 2020-02-22 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Design, Synthesis, and Structural Analysis of Cladosporin-Based Inhibitors of Malaria Parasites.
Acs Infect Dis., 7, 2021
|
|
4YE9
 
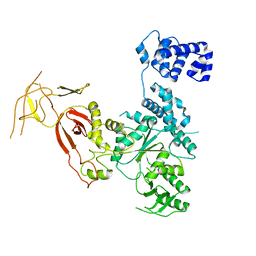 | |
1DT4
 
 | | CRYSTAL STRUCTURE OF NOVA-1 KH3 K-HOMOLOGY RNA-BINDING DOMAIN | | Descriptor: | NEURO-ONCOLOGICAL VENTRAL ANTIGEN 1 | | Authors: | Lewis, H.A, Chen, H, Edo, C, Buckanovich, R.J, Yang, Y.Y.L. | | Deposit date: | 2000-01-11 | | Release date: | 2000-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains.
Structure Fold.Des., 7, 1999
|
|
3ZO4
 
 | | The Synthesis and Evaluation of Diazaspirocyclic Protein Kinase Inhibitors | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-(4-PHENYL-1,9-DIAZASPIRO[5.5]UNDECAN-9-YL)-9H-PURINE, CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT ALPHA, ... | | Authors: | Allen, C.E, Chow, C.L, Caldwell, J.J, Westwood, I.M, van Montfort, R.L, Collins, I. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Synthesis and evaluation of heteroaryl substituted diazaspirocycles as scaffolds to probe the ATP-binding site of protein kinases.
Bioorg. Med. Chem., 21, 2013
|
|
2OJW
 
 | | Crystal structure of human glutamine synthetase in complex with ADP and phosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-15 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design.
J.Mol.Biol., 375, 2008
|
|
1MRG
 
 | | STUDIES ON CRYSTAL STRUCTURES ACTIVE CENTER GEOMETRY AND DEPURINE MECHANISM OF TWO RIBOSOME-INACTIVATING PROTEINS | | Descriptor: | ADENOSINE, ALPHA-MOMORCHARIN | | Authors: | Huang, Q, Liu, S, Tang, Y, Jin, S, Wang, Y. | | Deposit date: | 1994-07-01 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Studies on crystal structures, active-centre geometry and depurinating mechanism of two ribosome-inactivating proteins.
Biochem.J., 309, 1995
|
|
