3Q8X
 
 | | Structure of a toxin-antitoxin system bound to its substrate | | Descriptor: | Antidote of epsilon-zeta postsegregational killing system, SULFATE ION, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, ... | | Authors: | Mutschler, H, Gebhardt, M, Shoeman, R.L, Meinhart, A. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A novel mechanism of programmed cell death in bacteria by toxin-antitoxin systems corrupts peptidoglycan synthesis.
Plos Biol., 9, 2011
|
|
3U9E
 
 | | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA. | | Descriptor: | ARGININE, CHLORIDE ION, COENZYME A, ... | | Authors: | Tan, K, Zhou, M, Peterson, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-18 | | Release date: | 2011-11-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The crystal structure of a possible phosphate acetyl/butaryl transferase (from Listeria monocytogenes EGD-e) in complex with CoA.
To be Published
|
|
4KZC
 
 | | Structure of PI3K gamma with Imidazopyridine inhibitors | | Descriptor: | N-{6-[6-amino-5-(trifluoromethyl)pyridin-3-yl]imidazo[1,2-a]pyridin-2-yl}acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, E.A. | | Deposit date: | 2013-05-29 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure guided optimization of a fragment hit to imidazopyridine inhibitors of PI3K.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1JKM
 
 | | BREFELDIN A ESTERASE, A BACTERIAL HOMOLOGUE OF HUMAN HORMONE SENSITIVE LIPASE | | Descriptor: | BREFELDIN A ESTERASE | | Authors: | Wei, Y, Contreras, A.J, Sheffield, P, Osterlund, T, Derewenda, U, Matern, U.O, Derewenda, Z.S. | | Deposit date: | 1998-02-04 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of brefeldin A esterase, a bacterial homolog of the mammalian hormone-sensitive lipase.
Nat.Struct.Biol., 6, 1999
|
|
4KVN
 
 | | Crystal structure of Fab 39.29 in complex with Influenza Hemagglutinin A/Perth/16/2009 (H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | Fong, R, Swem, L.R, Lupardus, P.J. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Novel In vivo Human Plasmablast Enrichment Technique Allows Rapid Identification of Therapeutic Anti-Influenza A Antibodies
Cell Host Microbe, 14, 2013
|
|
1TMB
 
 | | MOLECULAR BASIS FOR THE INHIBITION OF HUMAN ALPHA-THROMBIN BY THE MACROCYCLIC PEPTIDE CYCLOTHEONAMIDE A | | Descriptor: | ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), HIRUGEN, ... | | Authors: | Qiu, X, Padmanabhan, K.P, Maryanoff, B.E, Tulinsky, A. | | Deposit date: | 1993-05-27 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis for the inhibition of human alpha-thrombin by the macrocyclic peptide cyclotheonamide A.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
1VY7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1VY6
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1M4L
 
 | | STRUCTURE OF NATIVE CARBOXYPEPTIDASE A AT 1.25 RESOLUTION | | Descriptor: | CARBOXYPEPTIDASE A, ZINC ION | | Authors: | Kilshtain-Vardi, A, Glick, M, Greenblatt, H.M, Goldblum, A, Shoham, G. | | Deposit date: | 2002-07-03 | | Release date: | 2003-01-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Refined structure of bovine carboxypeptidase A at 1.25 A resolution.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1ESF
 
 | | STAPHYLOCOCCAL ENTEROTOXIN A | | Descriptor: | CADMIUM ION, STAPHYLOCOCCAL ENTEROTOXIN A | | Authors: | Schad, E.M, Svensson, L.A. | | Deposit date: | 1995-05-25 | | Release date: | 1996-07-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superantigen staphylococcal enterotoxin type A.
EMBO J., 14, 1995
|
|
3V6E
 
 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
2YET
 
 | | Thermoascus GH61 isozyme A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYL GROUP, COPPER (II) ION, ... | | Authors: | Otten, H, Quinlan, R.J, Sweeney, M.D, Poulsen, J.-C.N, Johansen, K.S, Krogh, K.B.R.M, Joergensen, C.I, Tovborg, M, Anthonsen, A, Tryfona, T, Walter, C.P, Dupree, P, Xu, F, Davies, G.J, Walton, P.H, Lo Leggio, L. | | Deposit date: | 2011-03-30 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | Insights Into the Oxidative Degradation of Cellulose by a Copper Metalloenzyme that Exploits Biomass Components.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1SJF
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Cobalt Hexammine solution | | Descriptor: | COBALT HEXAMMINE(III), Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A conformational switch controls hepatitis delta virus ribozyme catalysis.
Nature, 429, 2004
|
|
4LUN
 
 | | Structure of the N-terminal mIF4G domain from S. cerevisiae Upf2, a protein involved in the degradation of mRNAs containing premature stop codons | | Descriptor: | CHLORIDE ION, Nonsense-mediated mRNA decay protein 2 | | Authors: | Fourati, Z, Roy, B, Millan, C, Courreux, P.D, Kervestin, S, van Tilbeurgh, H, He, F, Uson, I, Jacobson, A, Graille, M. | | Deposit date: | 2013-07-25 | | Release date: | 2014-07-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | A highly conserved region essential for NMD in the Upf2 N-terminal domain.
J.Mol.Biol., 426, 2014
|
|
2BXR
 
 | | Human Monoamine Oxidase A in complex with Clorgyline, Crystal Form A | | Descriptor: | AMINE OXIDASE [FLAVIN-CONTAINING] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | De Colibus, L, Binda, C, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2005-07-27 | | Release date: | 2005-08-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-Dimensional Structure of Human Monoamine Oxidase a (Mao A): Relation to the Structures of Rat Mao a and Human Mao B
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1U67
 
 | | Crystal Structure of Arachidonic Acid Bound to a Mutant of Prostagladin H Synthase-1 that Forms Predominantly 11-HPETE. | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARACHIDONIC ACID, PROTOPORPHYRIN IX CONTAINING CO, ... | | Authors: | Harman, C.A, Rieke, C.J, Garavito, R.M, Smith, W.L. | | Deposit date: | 2004-07-29 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of arachidonic Acid bound to a mutant of prostaglandin endoperoxide h synthase-1 that forms predominantly 11-hydroperoxyeicosatetraenoic Acid.
J.Biol.Chem., 279, 2004
|
|
2C69
 
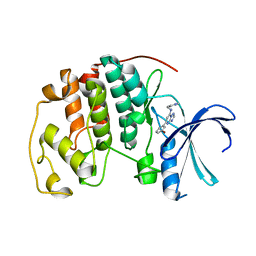 | | Crystal structure of the human CDK2 complexed with the triazolopyrimidine inhibitor | | Descriptor: | (5Z)-5-(3-BROMOCYCLOHEXA-2,5-DIEN-1-YLIDENE)-N-(PYRIDIN-4-YLMETHYL)-1,5-DIHYDRO[1,2,4]TRIAZOLO[1,5-A]PYRIMIDIN-7-AMINE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Richardson, C.M, Dokurno, P, Murray, J.B, Surgenor, A.E. | | Deposit date: | 2005-11-08 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Triazolo[1,5-a]pyrimidines as novel CDK2 inhibitors: protein structure-guided design and SAR.
Bioorg. Med. Chem. Lett., 16, 2006
|
|
2ISG
 
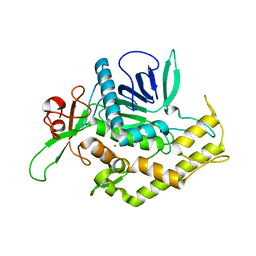 | | Botulinum Neurotoxin A Light Chain WT Crystal Form B | | Descriptor: | NICKEL (II) ION, Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 2024
|
|
8VSK
 
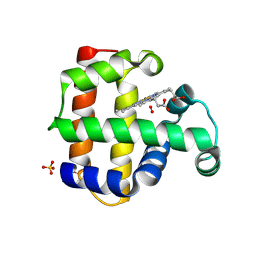 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 2024
|
|
8VKD
 
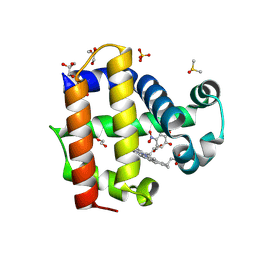 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 2024
|
|
3T9T
 
 | | Crystal structure of BTK mutant (F435T,K596R) complexed with Imidazo[1,5-a]quinoxaline | | Descriptor: | (2Z)-4-(dimethylamino)-N-{7-fluoro-4-[(2-methylphenyl)amino]imidazo[1,5-a]quinoxalin-8-yl}-N-methylbut-2-enamide, GLYCEROL, Tyrosine-protein kinase ITK/TSK | | Authors: | Han, S, Caspers, N. | | Deposit date: | 2011-08-03 | | Release date: | 2011-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imidazo[1,5-a]quinoxalines as irreversible BTK inhibitors for the treatment of rheumatoid arthritis.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1SJ3
 
 | | Hepatitis Delta Virus Gemonic Ribozyme Precursor, with Mg2+ Bound | | Descriptor: | MAGNESIUM ION, precursor form of the Hepatitis Delta virus ribozyme, small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H, Doudna, J.A. | | Deposit date: | 2004-03-02 | | Release date: | 2004-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme
catalysis
Nature, 429, 2004
|
|
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 2024
|
|
1S1S
 
 | | Crystal Structure of ZipA in complex with indoloquinolizin 10b | | Descriptor: | Cell division protein zipA, N-{3-[(12bS)-7-oxo-1,3,4,6,7,12b-hexahydroindolo[2,3-a]quinolizin-12(2H)-yl]propyl}propane-2-sulfonamide | | Authors: | Jennings, L.D, Foreman, K.W, Rush III, T.S, Tsao, D.H, Mosyak, L, Li, Y, Sukhdeo, M.N, Ding, W, Dushin, E.G, Kenny, C.H, Moghazeh, S.L, Petersen, P.J, Ruzin, A.V, Tuckman, M, Sutherland, A.G. | | Deposit date: | 2004-01-07 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of indolo[2,3-a]quinolizin-7-one inhibitors of the ZipA-FtsZ interaction
BIOORG.MED.CHEM.LETT., 14, 2004
|
|
