7ODK
 
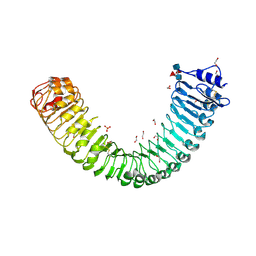 | | Plant peptide hormone receptor H1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roman, A.O, Jimenez-Sandoval, P, Santiago, J. | | Deposit date: | 2021-04-29 | | Release date: | 2022-02-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | HSL1 and BAM1/2 impact epidermal cell development by sensing distinct signaling peptides.
Nat Commun, 13, 2022
|
|
9M2Q
 
 | | Imidazole glycerol phosphate dehydratase from Mycobacterium tuberculosis, in complex with aminotriazole | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, Imidazoleglycerol-phosphate dehydratase, MANGANESE (II) ION | | Authors: | Raina, R, Kar, D, Singla, M, Tiwari, S, Kumari, S, Aneja, S, Kumar, V, Banerjee, S, Goyal, S, Pal, R.K, Vinothkumar, K.R, Biswal, B.K. | | Deposit date: | 2025-02-28 | | Release date: | 2025-06-18 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of Mycobacterium tuberculosis imidazole glycerol phosphate dehydratase in the apo state and in the presence of small molecules.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
9CCX
 
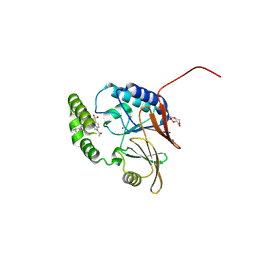 | | Crystal Structure of the Klebsiella pneumoniae LpxH/JH-LPH-86 complex | | Descriptor: | 1-(5-{4-[4-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, MANGANESE (II) ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
9CCY
 
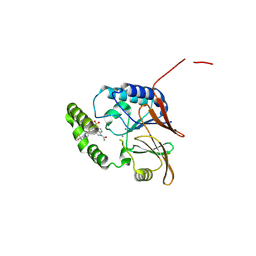 | | Crystal structure of the Klebsiella pneumoniae LpxH / JH-LPH-90 complex | | Descriptor: | 1-(5-{4-[6-(trifluoromethyl)pyridin-2-yl]piperazine-1-sulfonyl}-2,3-dihydro-1H-indol-1-yl)ethan-1-one, MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase | | Authors: | Cochrane, C.S, Zhou, P. | | Deposit date: | 2024-06-23 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Design and Evaluation of Pyridinyl Sulfonyl Piperazine LpxH Inhibitors with Potent Antibiotic Activity Against Enterobacterales.
Jacs Au, 4, 2024
|
|
8HBW
 
 | | Structure of human UCP1 in the ATP-bound state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, ... | | Authors: | Chen, L, Kang, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural basis for the binding of DNP and purine nucleotides onto UCP1.
Nature, 620, 2023
|
|
8HBV
 
 | | Structure of human UCP1 in the nucleotide-free state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CARDIOLIPIN, Mitochondrial brown fat uncoupling protein 1, ... | | Authors: | Chen, L, Kang, Y. | | Deposit date: | 2022-10-31 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Structural basis for the binding of DNP and purine nucleotides onto UCP1.
Nature, 620, 2023
|
|
2MU2
 
 | | NMR structure of the cap domain of NP_346487.1, a putative phosphoglycolate phosphatase from Streptococcus pneumoniae TIGR4 | | Descriptor: | Hydrolase, haloacid dehalogenase-like family | | Authors: | Jaudzems, K, Serrano, P, Pedrini, B, Geralt, M, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | J-UNIO protocol used for NMR structure determination of the 206-residue protein NP_346487.1 from Streptococcus pneumoniae TIGR4.
J.Biomol.Nmr, 61, 2015
|
|
6NO8
 
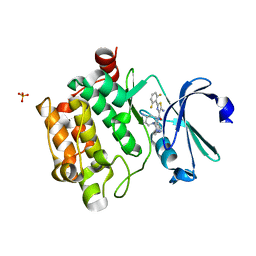 | |
1TSD
 
 | |
9D3W
 
 | | CRYSTAL STRUCTURE OF EGFR(L858R/T790M/C797S) IN COMPLEX WITH AUR-8250 | | Descriptor: | (4S)-1-(1,4-dioxaspiro[4.5]decan-8-yl)-N-{2-[(3S,4R)-3-fluoro-4-methoxypiperidin-1-yl]pyrimidin-4-yl}-2-methyl-1H-imidazo[1,2-b]pyrazol-6-amine, CHLORIDE ION, Epidermal growth factor receptor | | Authors: | Bell, J.A. | | Deposit date: | 2024-08-12 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Discovery of a Novel Mutant-Selective Epidermal Growth Factor Receptor Inhibitor Using an In Silico Enabled Drug Discovery Platform.
J.Med.Chem., 67, 2024
|
|
4EFM
 
 | | Crystal structure of H-Ras G12V in complex with GppNHp (state 1) | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Muraoka, S, Shima, F, Araki, M, Inoue, T, Yoshimoto, A, Ijiri, Y, Seki, N, Tamura, A, Kumasaka, T, Yamamoto, M, Kataoka, T. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the state 1 conformations of the GTP-bound H-Ras protein and its oncogenic G12V and Q61L mutants
Febs Lett., 586, 2012
|
|
4YD6
 
 | |
8H9F
 
 | | Human ATP synthase state 1 subregion 3 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ATP synthase F(0) complex subunit B1, mitochondrial, ... | | Authors: | Lai, Y, Zhang, Y, Liu, F, Gao, Y, Gong, H, Rao, Z. | | Deposit date: | 2022-10-25 | | Release date: | 2023-08-30 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure of the human ATP synthase.
Mol.Cell, 83, 2023
|
|
2NAV
 
 | | NMR solution structure of Ex-4[1-16]/pl14a | | Descriptor: | Exendin-4, Alpha/kappa-conotoxin pl14a chimera | | Authors: | Schroeder, C.I, Swedberg, J.E, Craik, D.J. | | Deposit date: | 2016-01-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-11-27 | | Method: | SOLUTION NMR | | Cite: | Truncated Glucagon-like Peptide-1 and Exendin-4 alpha-Conotoxin pl14a Peptide Chimeras Maintain Potency and alpha-Helicity and Reveal Interactions Vital for cAMP Signaling in Vitro.
J.Biol.Chem., 291, 2016
|
|
4ETQ
 
 | |
1TTA
 
 | |
1U2Y
 
 | | In situ extension as an approach for identifying novel alpha-amylase inhibitors, structure containing D-gluconhydroximo-1,5-lactam | | Descriptor: | (2S,3S,4R,5R)-6-(HYDROXYAMINO)-2-(HYDROXYMETHYL)-2,3,4,5-TETRAHYDROPYRIDINE-3,4,5-TRIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Numao, S, Li, C, Damager, I, Wrodnigg, T.M, Begum, A, Overall, C.M, Brayer, G.D, Withers, S.G. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | In Situ Extension as an Approach for Identifying Novel alpha-Amylase Inhibitors.
J.Biol.Chem., 279, 2004
|
|
2NBI
 
 | | Structure of the PSCD-region of the cell wall protein pleuralin-1 | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Rainer, D, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, H.R. | | Deposit date: | 2016-02-23 | | Release date: | 2016-12-21 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
9KYH
 
 | | Crystal structure of E.coli ac4C amidohydrolase YqfB in complex with ac4C | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, MAGNESIUM ION, N(4)-acetylcytidine amidohydrolase, ... | | Authors: | Guo, W.T, Meng, C.Y, Wen, Y, Wu, B.X. | | Deposit date: | 2024-12-09 | | Release date: | 2025-08-20 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural analysis of ASCH domain-containing proteins and their implications for nucleotide processing.
Structure, 2025
|
|
6NO9
 
 | | PIM1 in complex with Cpd16 (5-amino-N-(5-((4R,5R)-4-amino-5-fluoroazepan-1-yl)-1-methyl-1H-pyrazol-4-yl)-2-(2,6-difluorophenyl)thiazole-4-carboxamide) | | Descriptor: | 5-amino-N-{5-[(4R,5R)-4-amino-5-fluoroazepan-1-yl]-1-methyl-1H-pyrazol-4-yl}-2-(2,6-difluorophenyl)-1,3-thiazole-4-carboxamide, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Murray, J.M, Noland, C. | | Deposit date: | 2019-01-15 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Optimization of Pan-Pim Kinase Activity and Oral Bioavailability Leading to Diaminopyrazole (GDC-0339) for the Treatment of Multiple Myeloma.
J. Med. Chem., 62, 2019
|
|
4Y53
 
 | | Endothiapepsin in complex with fragment 54 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-difluorophenyl)imidazo[2,1-c][1,2,4]triazine, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-02-11 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
1U1F
 
 | | Structure of e. coli uridine phosphorylase complexed to 5-(m-(benzyloxy)benzyl)acyclouridine (BBAU) | | Descriptor: | 1-((2-HYDROXYETHOXY)METHYL)-5-(3-(BENZYLOXY)BENZYL)PYRIMIDINE-2,4(1H,3H)-DIONE, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Bu, W, Settembre, E.C, Ealick, S.E. | | Deposit date: | 2004-07-15 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for inhibition of Escherichia coli uridine phosphorylase by 5-substituted acyclouridines.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
9JTT
 
 | | Crystal Structure of Beta-glucosidase from the Indigo-producing Plant Polygonum tinctorium | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Yoneda, K, Himeno, M, Sakuraba, H, Ohshima, T. | | Deposit date: | 2024-10-07 | | Release date: | 2025-08-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Beta-glucosidase from the Indigo-producing Plant Polygonum tinctorium
To Be Published
|
|
1TTC
 
 | |
1U60
 
 | | MCSG APC5046 Probable glutaminase ybaS | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Probable glutaminase ybaS | | Authors: | Chang, C, Cuff, M.E, Joachimiak, A, Savchenko, A, Edwards, A, Skarina, T, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-07-28 | | Release date: | 2004-09-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Functional and structural characterization of four glutaminases from Escherichia coli and Bacillus subtilis.
Biochemistry, 47, 2008
|
|
