7BJD
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 3 | | Descriptor: | 2-methyl-2-(3-methyl-4-{[4-(methylamino)-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1H-pyrazol-1-yl)propanenitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJH
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 8 | | Descriptor: | CHLORIDE ION, N,N-dimethyl-7H-purin-6-amine, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJM
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 10 | | Descriptor: | 4-amino-6-{[(2-fluorophenyl)methyl]amino}-7-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK3
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 45 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3S)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJR
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 18 | | Descriptor: | 4-amino-7-methyl-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJJ
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 9 | | Descriptor: | 1H-pyrazolo[3,4-d]pyrimidin-4-amine, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJO
 
 | | Crystal structure of CHK1-10pt-mutant complex with compound 13 | | Descriptor: | 4-amino-6-(2,3-difluorophenyl)-7-methyl-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BJE
 
 | | Crystal structure of CHK1-10pt-mutant complex with adenine | | Descriptor: | ADENINE, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-14 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
7BK2
 
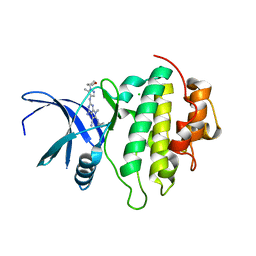 | | Crystal structure of CHK1-10pt-mutant complex with compound 44 | | Descriptor: | 4-amino-7-methyl-2-({5-methyl-1-[(3R)-oxolan-3-yl]-1H-pyrazol-4-yl}amino)-6-[(2R)-2-methylpyrrolidin-1-yl]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, CHLORIDE ION, Serine/threonine-protein kinase Chk1 | | Authors: | Dokurno, P, Surgenor, A.E, Williamson, D.S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and Synthesis of Pyrrolo[2,3- d ]pyrimidine-Derived Leucine-Rich Repeat Kinase 2 (LRRK2) Inhibitors Using a Checkpoint Kinase 1 (CHK1)-Derived Crystallographic Surrogate.
J.Med.Chem., 64, 2021
|
|
4N2H
 
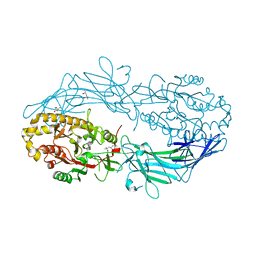 | | Crystal structure of Protein Arginine Deiminase 2 (D177A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Protein-arginine deiminase type-2 | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.808 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4NA9
 
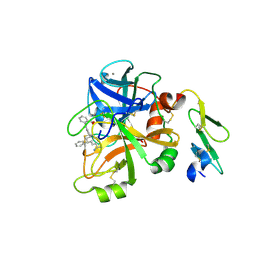 | | Factor VIIa in complex with the inhibitor 3'-amino-5'-[(2s,4r)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid | | Descriptor: | 3'-amino-5'-[(2S,4R)-6-carbamimidoyl-4-phenyl-1,2,3,4-tetrahydroquinolin-2-yl]biphenyl-2-carboxylic acid, CALCIUM ION, Coagulation factor VII heavy chain, ... | | Authors: | Wei, A. | | Deposit date: | 2013-10-21 | | Release date: | 2014-02-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Tetrahydroquinoline Derivatives as Potent and Selective Factor XIa Inhibitors.
J.Med.Chem., 57, 2014
|
|
4MQD
 
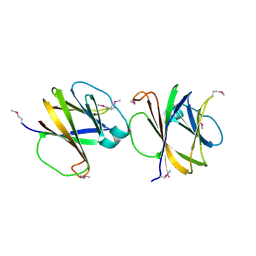 | | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis | | Descriptor: | DNA-entry nuclease inhibitor | | Authors: | Chang, C, Mack, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-16 | | Release date: | 2013-10-09 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of ComJ, inhibitor of the DNA degrading activity of NucA, from Bacillus subtilis
To be Published
|
|
4ERD
 
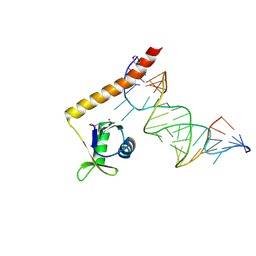 | | Crystal structure of the C-terminal domain of Tetrahymena telomerase protein p65 in complex with stem IV of telomerase RNA | | Descriptor: | 5'-R(P*GP*GP*UP*CP*GP*AP*CP*AP*UP*CP*UP*UP*CP*GP*GP*AP*UP*GP*GP*AP*CP*C)-3', POTASSIUM ION, Telomerase associated protein p65 | | Authors: | Singh, M, Wang, Z, Koo, B.-K, Patel, A, Cascio, D, Collins, K, Feigon, J. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-20 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.589 Å) | | Cite: | Structural Basis for Telomerase RNA Recognition and RNP Assembly by the Holoenzyme La Family Protein p65.
Mol.Cell, 47, 2012
|
|
7RTA
 
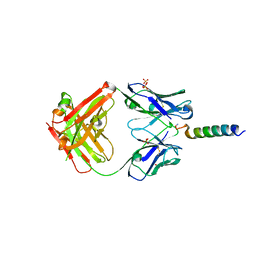 | | Crystal structures of human PYY and NPY | | Descriptor: | 4A3B2-B Fab heavy chain, 4A3B2-B Fab light chain, Neuropeptide Y, ... | | Authors: | Langley, D.B, Christ, D. | | Deposit date: | 2021-08-12 | | Release date: | 2022-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structures of human neuropeptide Y (NPY) and peptide YY (PYY).
Neuropeptides, 92, 2022
|
|
4ESS
 
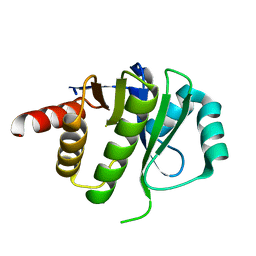 | | Crystal Structure of E6D/L155R variant of de novo designed serine hydrolase OSH55, Northeast Structural Genomics Consortium (NESG) Target OR187 | | Descriptor: | OR187 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Kornhaber, K, Kornhaber, G, Rajagopalan, S, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-04-23 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9971 Å) | | Cite: | Design of activated serine-containing catalytic triads with atomic-level accuracy.
Nat.Chem.Biol., 10, 2014
|
|
4EUF
 
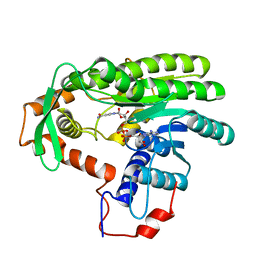 | | Crystal structure of Clostridium acetobutulicum trans-2-enoyl-CoA reductase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative reductase CA_C0462, SODIUM ION | | Authors: | Hu, K, Zhao, M, Zhang, T, Yang, S, Ding, J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-11-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of trans-2-enoyl-CoA reductases from Clostridium acetobutylicum and Treponema denticola: insights into the substrate specificity and the catalytic mechanism
Biochem.J., 449, 2013
|
|
4N2F
 
 | | Crystal structure of Protein Arginine Deiminase 2 (D169A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4N24
 
 | | Crystal structure of Protein Arginine Deiminase 2 (100 uM Ca2+) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-04 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
7BHF
 
 | | DARPin_D5/Her3 domain 4 complex, orthorhombic crystals | | Descriptor: | ACETATE ION, DARPin_D5, Isoform 4 of Receptor tyrosine-protein kinase erbB-3 | | Authors: | Mittl, P.R.E, Radom, F, Pluckthun, A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Crystal structures of HER3 extracellular domain 4 in complex with the designed ankyrin-repeat protein D5.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
4N2M
 
 | | Crystal structure of Protein Arginine Deiminase 2 (E354A, 0 mM Ca2+) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Slade, D.J, Zhang, X, Fang, P, Dreyton, C.J, Zhang, Y, Gross, M.L, Guo, M, Coonrod, S.A, Thompson, P.R. | | Deposit date: | 2013-10-05 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Protein arginine deiminase 2 binds calcium in an ordered fashion: implications for inhibitor design.
Acs Chem.Biol., 10, 2015
|
|
4E69
 
 | | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure | | Descriptor: | 1,2-ETHANEDIOL, 2-dehydro-3-deoxygluconokinase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-03-15 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a sugar kinase (target EFI-502132) from Oceanicola granulosus, unliganded structure
To be Published
|
|
4EDN
 
 | | Crystal structure of beta-parvin CH2 domain in complex with paxillin LD1 motif | | Descriptor: | Beta-parvin, Paxillin, SULFATE ION | | Authors: | Stiegler, A.L, Draheim, K.M, Li, X, Chayen, N.E, Calderwood, D.A, Boggon, T.J. | | Deposit date: | 2012-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for paxillin binding and focal adhesion targeting of beta-parvin.
J.Biol.Chem., 287, 2012
|
|
4K37
 
 | | Native anSMEcpe with bound AdoMet | | Descriptor: | Anaerobic sulfatase-maturating enzyme, CHLORIDE ION, GLYCEROL, ... | | Authors: | Goldman, P.J, Drennan, C.L. | | Deposit date: | 2013-04-10 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KF1
 
 | | Crystal structure of SsoPox W263I in complex with C10HTL | | Descriptor: | (4S)-4-(decanoylamino)-5-hydroxy-3,4-dihydro-2H-thiophenium, 1,2-ETHANEDIOL, Aryldialkylphosphatase, ... | | Authors: | Gotthard, G, Hiblot, J, Chabriere, E, Elias, M. | | Deposit date: | 2013-04-26 | | Release date: | 2013-10-02 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Active Site Loop Conformations Mediate Promiscuous Activities in the Lactonase SsoPox.
Plos One, 8, 2013
|
|
4EIR
 
 | | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases (PMO-2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Li, X, Beeson, W.T, Phillips, C.M, Marletta, M.A, Cate, J.H. | | Deposit date: | 2012-04-05 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases.
Structure, 20, 2012
|
|
