3BJK
 
 | | Crystal structure of HI0827, a hexameric broad specificity acyl-coenzyme A thioesterase: The Asp44Ala mutant enzyme | | Descriptor: | 1,2-ETHANEDIOL, Acyl-CoA thioester hydrolase HI0827, CITRIC ACID | | Authors: | Willis, M.A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of YciA from Haemophilus influenzae (HI0827), a Hexameric Broad Specificity Acyl-Coenzyme A Thioesterase.
Biochemistry, 47, 2008
|
|
3B04
 
 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with oIPP. | | Descriptor: | 1-deoxy-1-[(4aR)-4a-[(2R)-1-hydroxy-5-{[(S)-hydroxy(phosphonooxy)phosphoryl]oxy}-3-methylidenepentan-2-yl]-7,8-dimethyl-2,4-dioxo-3,4,4a,5-tetrahydrobenzo[g]pteridin-10(2H)-yl]-5-O-phosphono-D-ribitol, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION | | Authors: | Unno, H, Nagai, T, Hemmi, H. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Covalent modification of reduced flavin mononucleotide in type-2 isopentenyl diphosphate isomerase by active-site-directed inhibitors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B4N
 
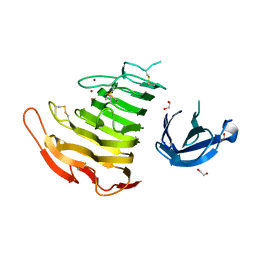 | | Crystal Structure Analysis of Pectate Lyase PelI from Erwinia chrysanthemi | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Endo-pectate lyase, ... | | Authors: | Creze, C, Castang, S, Derivery, E, Haser, R, Shevchik, V, Gouet, P. | | Deposit date: | 2007-10-24 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The Crystal Structure of Pectate Lyase PelI from Soft Rot Pathogen Erwinia chrysanthemi in Complex with Its Substrate.
J.Biol.Chem., 283, 2008
|
|
6P38
 
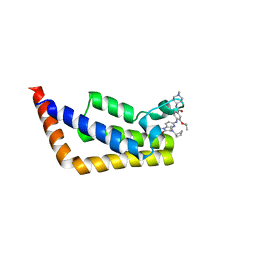 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-4-cyclopentyl-1,3-dimethyl-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-N-(1-methylpiperidin-4-yl)-3-[(propan-2-yl)oxy]benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dual Inhibition of TAF1 and BET Bromodomains from the BI-2536 Kinase Inhibitor Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
8PO2
 
 | |
7LHF
 
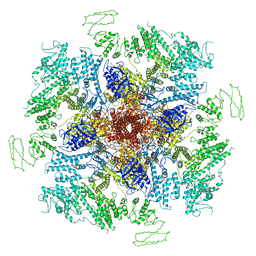 | | Structure of full-length IP3R1 channel solubilized in LNMG & lipid in the apo-state | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, Inositol 1,4,5-trisphosphate receptor type 1, ZINC ION | | Authors: | Baker, M.R, Fan, G, Baker, M.L, Serysheva, I.I. | | Deposit date: | 2021-01-22 | | Release date: | 2021-06-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Cryo-EM structure of type 1 IP3R channel in a lipid bilayer
Commun Biol, 4, 2021
|
|
3BFT
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | Descriptor: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2007-11-23 | | Release date: | 2008-10-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
7L6X
 
 | | Crystal structure of the tandem bromodomain (BD1, BD2) of human TAF1 bound to GNE-371 | | Descriptor: | 1,2-ETHANEDIOL, 6-(but-3-en-1-yl)-4-[1-methyl-6-(morpholine-4-carbonyl)-1H-benzimidazol-4-yl]-1,6-dihydro-7H-pyrrolo[2,3-c]pyridin-7-one, Transcription initiation factor TFIID subunit 1 | | Authors: | Karim, M.R, Schonbrunn, E. | | Deposit date: | 2020-12-24 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Dual TAF1-ATR Inhibitors and Ligand-Induced Structural Changes of the TAF1 Tandem Bromodomain.
J.Med.Chem., 65, 2022
|
|
7L9K
 
 | | Crystal structure of the second bromodomain (BD2) of human BRD2 bound to LRRK2-IN-1 | | Descriptor: | 1,4-BUTANEDIOL, 2-[(2-methoxy-4-{[4-(4-methylpiperazin-1-yl)piperidin-1-yl]carbonyl}phenyl)amino]-5,11-dimethyl-5,11-dihydro-6H-pyrimido[4,5-b][1,4]benzodiazepin-6-one, Bromodomain-containing protein 2 | | Authors: | Karim, M.R, Bikowitz, M.J, Schonbrunn, E. | | Deposit date: | 2021-01-04 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential BET Bromodomain Inhibition by Dihydropteridinone and Pyrimidodiazepinone Kinase Inhibitors.
J.Med.Chem., 64, 2021
|
|
8PO4
 
 | |
7LB1
 
 | |
8BI5
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0737 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-~{N}-methyl-~{N}-(1-propan-2-ylpiperidin-4-yl)-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0737
To Be Published
|
|
8PNZ
 
 | |
8BI6
 
 | | Crystal structure of human Choline Kinase A in complex with UNC0638 | | Descriptor: | 1,2-ETHANEDIOL, 2-cyclohexyl-6-methoxy-N-[1-(1-methylethyl)piperidin-4-yl]-7-(3-pyrrolidin-1-ylpropoxy)quinazolin-4-amine, CHLORIDE ION, ... | | Authors: | Diaz-Saez, L, Ward, J, Kennedy, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-01 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human Choline Kinase A in complex with UNC0638
To Be Published
|
|
7L1Z
 
 | | Unlocking the structural features for the exo-xylobiosidase activity of an unusual GH11 member identified in a compost-derived consortium - NT-truncated form | | Descriptor: | 1,2-ETHANEDIOL, Exo-B-1,4-beta-xylanase, SULFATE ION | | Authors: | Kadowaki, M.A.S, Polikarpov, I, Briganti, L, Evangelista, D.E. | | Deposit date: | 2020-12-15 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Unlocking the structural features for the xylobiohydrolase activity of an unusual GH11 member identified in a compost-derived consortium.
Biotechnol.Bioeng., 118, 2021
|
|
3B06
 
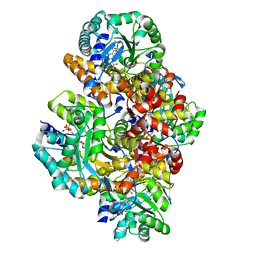 | | Crystal structure of Sulfolobus shibatae isopentenyl diphosphate isomerase in complex with reduced FMN and DMAPP. | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, DIMETHYLALLYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, ... | | Authors: | Unno, H, Nagai, T, Hemmi, H. | | Deposit date: | 2011-06-03 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Covalent modification of reduced flavin mononucleotide in type-2 isopentenyl diphosphate isomerase by active-site-directed inhibitors.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8BK3
 
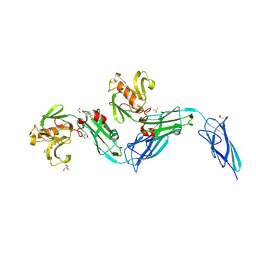 | | Crystal structure of the transpeptidase LdtMt2 from Mycobacterium tuberculosis in complex with diepoxide ketone 1 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2~{S},3~{R})-3-phenyloxiran-2-yl]ethanone, DIMETHYL SULFOXIDE, ... | | Authors: | de Munnik, M, Schofield, C.J. | | Deposit date: | 2022-11-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | alpha beta , alpha ' beta '-Diepoxyketones are mechanism-based inhibitors of nucleophilic cysteine enzymes.
Chem.Commun.(Camb.), 59, 2023
|
|
6PAL
 
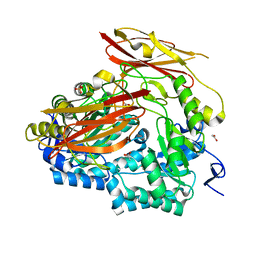 | | Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus | | Descriptor: | ACETATE ION, SULFATE ION, Uncharacterized protein | | Authors: | Tamura, K, Brumer, H, van Petegem, F. | | Deposit date: | 2019-06-11 | | Release date: | 2020-04-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.818 Å) | | Cite: | Synergy between Cell Surface Glycosidases and Glycan-Binding Proteins Dictates the Utilization of Specific Beta(1,3)-Glucans by Human GutBacteroides.
Mbio, 11, 2020
|
|
8BLJ
 
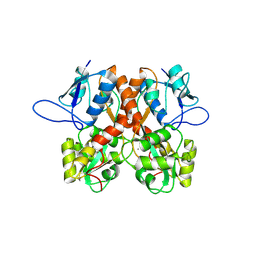 | | Crystal structure of the ligand-binding domain (LBD) of human iGluR Delta-1 (GluD1), apo state | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Heroven, C, Malinauskas, T, Aricescu, A.R. | | Deposit date: | 2022-11-09 | | Release date: | 2023-11-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | GluD1 binds GABA and controls inhibitory plasticity.
Science, 382, 2023
|
|
2PI2
 
 | | Full-length Replication protein A subunits RPA14 and RPA32 | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Replication protein A 14 kDa subunit, Replication protein A 32 kDa subunit | | Authors: | Deng, X, Borgstahl, G.E. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Full-length Human RPA14/32 Complex Gives Insights into the Mechanism of DNA Binding and Complex Formation.
J.Mol.Biol., 374, 2007
|
|
8GLP
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (Consensus LSU focused refined structure) | | Descriptor: | 1,4-DIAMINOBUTANE, 18S rRNA, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Watson, Z.L, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-03-22 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.67 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
2PN9
 
 | | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA modified aptamer | | Descriptor: | 5'-R(*GP*GP*AP*GP*CP*CP*UP*GP*GP*GP*AP*GP*CP*UP*CP*C)-3', RNA 16-mer with locked residues 9-10 | | Authors: | Lebars, I, Richard, T, Di Primo, C, Toulme, J.-J. | | Deposit date: | 2007-04-24 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a kissing complex formed between the TAR RNA element of HIV-1 and a LNA-modified aptamer
Nucleic Acids Res., 35, 2007
|
|
2Q9J
 
 | | Crystal structure of the C217S mutant of diaminopimelate epimerase | | Descriptor: | 1,2-ETHANEDIOL, Diaminopimelate epimerase, SULFATE ION | | Authors: | Pillai, B, Cherney, M, Diaper, C.M, Sutherland, A, Blanchard, J.S, Vederas, J.C. | | Deposit date: | 2007-06-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dynamics of catalysis revealed from the crystal structures of mutants of diaminopimelate epimerase.
Biochem.Biophys.Res.Commun., 363, 2007
|
|
8C3G
 
 | | Crystal structure of DYRK1A in complex with AZ191 | | Descriptor: | 1,2-ETHANEDIOL, Dual specificity tyrosine-phosphorylation-regulated kinase 1A, N-[2-methoxy-4-(4-methylpiperazin-1-yl)phenyl]-4-(1-methylpyrrolo[2,3-c]pyridin-3-yl)pyrimidin-2-amine, ... | | Authors: | Grygier, P, Pustelny, K, Dubin, G, Czarna, A. | | Deposit date: | 2022-12-23 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspective on the design of selective DYRK1B inhibitors
To Be Published
|
|
2PPY
 
 | | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Kavyashree, M, Sekar, K, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-01 | | Release date: | 2008-05-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of Enoyl-CoA hydrates (gk_1992) from Geobacillus Kaustophilus HTA426
To be Published
|
|
