6T3F
 
 | | Crystal structure Nipah virus fusion glycoprotein in complex with a neutralising Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab66 heavy chain, ... | | Authors: | Avanzato, V.A, Pryce, R, Walter, T.S, Bowden, T.A. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structural basis for antibody-mediated neutralization of Nipah virus reveals a site of vulnerability at the fusion glycoprotein apex.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6T4R
 
 | | Crystal structure of Trypanosoma brucei Morn1 | | Descriptor: | MORN repeat-containing protein 1 | | Authors: | Grishkovskaya, I, Kostan, J, Sajko, S, Morriswood, B, Djinovic-Carugo, K. | | Deposit date: | 2019-10-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Structures of three MORN repeat proteins and a re-evaluation of the proposed lipid-binding properties of MORN repeats.
Plos One, 15, 2020
|
|
1BVY
 
 | | COMPLEX OF THE HEME AND FMN-BINDING DOMAINS OF THE CYTOCHROME P450(BM-3) | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, PROTEIN (CYTOCHROME P450 BM-3), ... | | Authors: | Sevrioukova, I.F, Li, H, Zhang, H, Peterson, J.A, Poulos, T.L. | | Deposit date: | 1998-09-21 | | Release date: | 1999-02-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structure of a cytochrome P450-redox partner electron-transfer complex.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8HVP
 
 | | STRUCTURE AT 2.5-ANGSTROMS RESOLUTION OF CHEMICALLY SYNTHESIZED HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE COMPLEXED WITH A HYDROXYETHYLENE*-BASED INHIBITOR | | Descriptor: | HIV-1 PROTEASE, INHIBITOR VAL-SER-GLN-ASN-LEU-PSI(CH(OH)-CH2)-VAL-ILE-VAL (U-85548E) | | Authors: | Jaskolski, M, Miller, M, Tomasselli, A.G, Sawyer, T.K, Staples, D.G, Heinrikson, R.L, Schneider, J, Kent, S.B.H, Wlodawer, A. | | Deposit date: | 1990-10-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure at 2.5-A resolution of chemically synthesized human immunodeficiency virus type 1 protease complexed with a hydroxyethylene-based inhibitor.
Biochemistry, 30, 1991
|
|
2MUT
 
 | | Solution structure of the F231L mutant ERCC1-XPF dimerization region | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Faridounnia, M, Wienk, H, Kovacic, L, Folkers, G.E, Jaspers, N.G.J, Kaptein, R, Hoeijmakers, J.H.J, Boelens, R. | | Deposit date: | 2014-09-17 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Cerebro-oculo-facio-skeletal Syndrome Point Mutation F231L in the ERCC1 DNA Repair Protein Causes Dissociation of the ERCC1-XPF Complex.
J.Biol.Chem., 290, 2015
|
|
5JYG
 
 | |
6VK6
 
 | | Crystal Structure of Methylosinus trichosporium OB3b Soluble Methane Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Jones, J.C, Banerjee, R, Shi, K, Aihara, H, Lipscomb, J.D. | | Deposit date: | 2020-01-18 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural Studies of theMethylosinus trichosporiumOB3b Soluble Methane Monooxygenase Hydroxylase and Regulatory Component Complex Reveal a Transient Substrate Tunnel.
Biochemistry, 59, 2020
|
|
6JXM
 
 | | Crystal Structure of phi29 pRNA domain II | | Descriptor: | BARIUM ION, MAGNESIUM ION, RNA (97-mer) | | Authors: | Cai, R, Price, I.R, Ding, F, Wu, F, Chen, T, Zhang, Y, Liu, G, Jardine, P.J, Lu, C, Ke, A. | | Deposit date: | 2019-04-24 | | Release date: | 2019-08-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | ATP/ADP modulates gp16-pRNA conformational change in the Phi29 DNA packaging motor.
Nucleic Acids Res., 47, 2019
|
|
1XCK
 
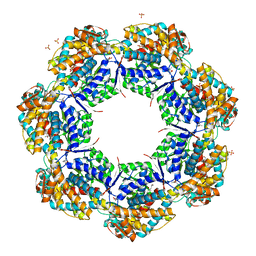 | | Crystal structure of apo GroEL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 60 kDa chaperonin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bartolucci, C, Lamba, D, Grazulis, S, Manakova, E, Heumann, H. | | Deposit date: | 2004-09-02 | | Release date: | 2005-10-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Crystal structure of wild-type chaperonin GroEL
J.Mol.Biol., 354, 2005
|
|
6MPB
 
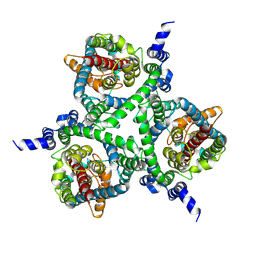 | | Cryo-EM structure of the human neutral amino acid transporter ASCT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMINE, Neutral amino acid transporter B(0) | | Authors: | Yu, X, Han, S. | | Deposit date: | 2018-10-05 | | Release date: | 2019-11-06 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures of the human glutamine transporter SLC1A5 (ASCT2) in the outward-facing conformation.
Elife, 8, 2019
|
|
8AV8
 
 | | Small molecular stabilizer for ERalpha and 14-3-3 (1075300) | | Descriptor: | 14-3-3 protein sigma, 2-chloranyl-~{N}-[[1-(1-phenoxycyclopentyl)carbonylpiperidin-4-yl]methyl]ethanamide, Estrogen receptor, ... | | Authors: | Visser, E.J, Vandenboorn, E.M.F, Ottmann, C. | | Deposit date: | 2022-08-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Optimization of Covalent, Small-Molecule Stabilizers of the 14-3-3 sigma /ER alpha Protein-Protein Interaction from Nonselective Fragments.
J.Am.Chem.Soc., 145, 2023
|
|
8H2H
 
 | | Cryo-EM structure of a Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | Group II intron-encoded protein LtrA, LtrB, RNA (5'-R(P*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*C)-3') | | Authors: | Liu, N, Dong, X.L, Qu, G.S, Wang, J, Wang, H.W, Belfort, M. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Functionalized graphene grids with various charges for single-particle cryo-EM.
Nat Commun, 13, 2022
|
|
5K3G
 
 | | Crystals structure of Acyl-CoA oxidase-1 in Caenorhabditis elegans, Apo form-I | | Descriptor: | Acyl-coenzyme A oxidase | | Authors: | Zhang, X, Li, K, Jones, R.A, Bruner, S.D, Butcher, R.A. | | Deposit date: | 2016-05-19 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.859 Å) | | Cite: | Structural characterization of acyl-CoA oxidases reveals a direct link between pheromone biosynthesis and metabolic state in Caenorhabditis elegans.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
8AMY
 
 | | High-resolution crystal structure of the Mu8.1 conotoxin from Conus Mucronatus | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICARBONATE ION, ... | | Authors: | Mueller, E, Hackney, C.M, Ellgaard, L, Morth, J.P. | | Deposit date: | 2022-08-04 | | Release date: | 2023-08-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-resolution crystal structure of the Mu8.1 conotoxin from Conus mucronatus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
1G6Q
 
 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
6C3O
 
 | | Cryo-EM structure of human KATP bound to ATP and ADP in quatrefoil form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Lee, K.P.K, Chen, J, MacKinnon, R. | | Deposit date: | 2018-01-10 | | Release date: | 2018-01-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular structure of human KATP in complex with ATP and ADP
Elife, 6, 2017
|
|
1XIW
 
 | | Crystal structure of human CD3-e/d dimer in complex with a UCHT1 single-chain antibody fragment | | Descriptor: | T-cell surface glycoprotein CD3 delta chain, T-cell surface glycoprotein CD3 epsilon chain, immunoglobulin heavy chain variable region, ... | | Authors: | Arnett, K.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
8HDD
 
 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
1XZY
 
 | | Solution structure of the P30-trans form of Alpha Hemoglobin Stabilizing Protein (AHSP) | | Descriptor: | Alpha-hemoglobin stabilizing protein | | Authors: | Gell, D.A, Feng, L, Zhou, S, Kong, Y, Lee, C, Weiss, M.J, Shi, Y, Mackay, J.P. | | Deposit date: | 2004-11-12 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Molecular mechanism of AHSP-mediated stabilization of alpha-hemoglobin
Cell(Cambridge,Mass.), 119, 2004
|
|
8T6I
 
 | | Structure of VHH-Fab complex with engineered Crystal Kappa region | | Descriptor: | Fab heavy chain, Fab light chain, GLYCEROL, ... | | Authors: | Filippova, E.V, Thompson, I, Kossiakoff, A.A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
5N2F
 
 | | Structure of PD-L1/small-molecule inhibitor complex | | Descriptor: | 4-[[4-[[3-(2,3-dihydro-1,4-benzodioxin-6-yl)-2-methyl-phenyl]methoxy]-2,5-bis(fluoranyl)phenyl]methylamino]-3-oxidanylidene-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Guzik, K, Zak, K.M, Grudnik, P, Dubin, G, Holak, T.A. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small-Molecule Inhibitors of the Programmed Cell Death-1/Programmed Death-Ligand 1 (PD-1/PD-L1) Interaction via Transiently Induced Protein States and Dimerization of PD-L1.
J. Med. Chem., 60, 2017
|
|
8AU4
 
 | | Structural insights reveal a heterotetramer between oncogenic K-Ras4BG12V and Rgl2, a RalA/B activator | | Descriptor: | Ral guanine nucleotide dissociation stimulator-like 2 | | Authors: | Tariq, M, Ikeya, T, Togashi, N, Fairall, L, Alejo, C.B, Kamei, S, Alonso, B.R, Campillo, M.A.M, Hudson, A, Ito, Y, Schwabe, J, Dominguez, C, Tanaka, K. | | Deposit date: | 2022-08-25 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-25 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the complex of oncogenic KRas4B G12V and Rgl2, a RalA/B activator.
Life Sci Alliance, 7, 2024
|
|
1BSN
 
 | |
8SXS
 
 | |
6MZB
 
 | | Cryo-EM structure of phosphodiesterase 6 | | Descriptor: | GUANOSINE-3',5'-MONOPHOSPHATE, MAGNESIUM ION, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit gamma, ... | | Authors: | Gulati, S, Palczewski, K. | | Deposit date: | 2018-11-04 | | Release date: | 2019-03-06 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of phosphodiesterase 6 reveals insights into the allosteric regulation of type I phosphodiesterases.
Sci Adv, 5, 2019
|
|
