6CPO
 
 | | Crystal structure of DR15 presenting the RQ13 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Farenc, C, Gras, S, Rossjohn, J. | | Deposit date: | 2018-03-13 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CD4+T cell-mediated HLA class II cross-restriction in HIV controllers.
Sci Immunol, 3, 2018
|
|
5GW8
 
 | | Crystal structure of a putative DAG-like lipase (MgMDL2) from Malassezia globosa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Xu, J, Xu, H, Liu, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Malassezia globosa MgMDL2 lipase: Crystal structure and rational modification of substrate specificity.
Biochem. Biophys. Res. Commun., 488, 2017
|
|
7L17
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Melibiose carrier protein | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
5UOP
 
 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 18) | | Descriptor: | (1S,2S,5R)-8'-[(3-chloro-4-fluorophenyl)methyl]-2'-[2-(2,5-dioxo-2,5-dihydro-1H-pyrrol-1-yl)ethyl]-6'-hydroxy-9',10'-dihydro-2'H-spiro[bicyclo[3.1.0]hexane-2,3'-imidazo[5,1-a][2,6]naphthyridine]-1',5',7'(8'H)-trione, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8AGF
 
 | |
6PZS
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with JR005 | | Descriptor: | 4-[({[(1R,2R,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl]methyl}amino)methyl]-N-hydroxybenzamide, CHLORIDE ION, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-08-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Exploring Structural Determinants of Inhibitor Affinity and Selectivity in Complexes with Histone Deacetylase 6.
J.Med.Chem., 63, 2020
|
|
8SU6
 
 | |
6YKK
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.236 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
8AH2
 
 | | Crystal structure of human 14-3-3 zeta fused to the NPM1 peptide including phosphoserine-48 | | Descriptor: | 14-3-3 protein zeta/delta,Nucleophosmin | | Authors: | Boyko, K.M, Kapitonova, A.A, Tugaeva, K.V, Varfolomeeva, L.A, Sluchanko, N.N. | | Deposit date: | 2022-07-20 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the recognition by 14-3-3 proteins of a conditional binding site within the oligomerization domain of human nucleophosmin.
Biochem.Biophys.Res.Commun., 627, 2022
|
|
7QCQ
 
 | | VHH Z70 in interaction with PHF6 Tau peptide | | Descriptor: | GLYCEROL, Tau 301-312, VHH Z70 | | Authors: | Danis, C, Dupre, E, Zejneli, O, Caillierez, R, Arrial, A, Begard, S, Loyens, A, Mortelecque, J, Cantrelle, F.-X, Hanoulle, X, Rain, J.-C, Colin, M, Buee, L, Landrieu, I. | | Deposit date: | 2021-11-25 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Tau seeding by targeting Tau nucleation core within neurons with a single domain antibody fragment.
Mol.Ther., 30, 2022
|
|
7QB7
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345 and L-Proline | | Descriptor: | 1,2-ETHANEDIOL, PROLINE, Proline--tRNA ligase, ... | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-18 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT345
To Be Published
|
|
6JGC
 
 | | Crystal structure of barley exohydrolaseI W286Y mutant in complex with glucose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-13 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6FBX
 
 | |
8SKH
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent pyrazoline based inhibitors | | Descriptor: | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase nsp5 | | Authors: | Knapp, M.S, Ornelas, E. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Discovery of Potent Pyrazoline-Based Covalent SARS-CoV-2 Main Protease Inhibitors.
Chembiochem, 24, 2023
|
|
6YKV
 
 | | Neisseria gonorrhoeae Leucyl-tRNA Synthetase in Complex with Compound 11g | | Descriptor: | 1,2-ETHANEDIOL, Leucine--tRNA ligase, MAGNESIUM ION, ... | | Authors: | Pang, L, Strelkov, S.V, Weeks, S.D. | | Deposit date: | 2020-04-06 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Synthesis and structure-activity studies of novel anhydrohexitol-based Leucyl-tRNA synthetase inhibitors.
Eur.J.Med.Chem., 211, 2021
|
|
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8ALP
 
 | | Botulinum neurotoxin A6 cell binding domain crystal form II | | Descriptor: | 1,2-ETHANEDIOL, Bont/A1, DI(HYDROXYETHYL)ETHER | | Authors: | Gregory, K.S, Acharya, K.R, Liu, S.M. | | Deposit date: | 2022-08-01 | | Release date: | 2022-09-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of the Clostridium botulinum Neurotoxin A6 Cell Binding Domain Alone and in Complex with GD1a Reveal Significant Conformational Flexibility.
Int J Mol Sci, 23, 2022
|
|
6CQH
 
 | | Crystal Structure of the Human vaccinia-related kinase bound to a N-propynyl-N-ethyl-dihydropteridine inhibitor | | Descriptor: | (7R)-2-[(3,5-difluoro-4-hydroxyphenyl)amino]-8-ethyl-7-methyl-5-(prop-2-yn-1-yl)-7,8-dihydropteridin-6(5H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | dos Reis, C.V, de Souza, G.P, Counago, R.M, Azevedo, A, Guimaraes, C, Mascarello, A, Gama, F, Ferreira, M, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-03-15 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Human vaccinia-related kinase bound to a N-propynyl-N-ethyl-dihydropteridine inhibitor
To Be Published
|
|
6YLC
 
 | | Biochemical, Cellular and Structural Characterization of Novel ERK3 Inhibitors | | Descriptor: | 5-fluoranyl-2-[5-[[1-(1-methylpiperidin-4-yl)pyrazol-4-yl]amino]-[1,2,3]triazolo[4,5-d]pyrimidin-3-yl]benzenecarbonitrile, Mitogen-activated protein kinase 6 | | Authors: | Graedler, U. | | Deposit date: | 2020-04-07 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Biochemical, cellular and structural characterization of novel and selective ERK3 inhibitors.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
5GO5
 
 | | Structure of sortase E from Streptomyces avermitilis | | Descriptor: | GLYCINE, SULFATE ION, sortase | | Authors: | Das, S, Pawale, V.S, Dadireddy, V, Roy, R.P, Ramakumar, S. | | Deposit date: | 2016-07-26 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and specificity of a new class of Ca2+-independent housekeeping sortase from Streptomyces avermitilis provide insights into its non-canonical substrate preference.
J. Biol. Chem., 292, 2017
|
|
7QC1
 
 | | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436 | | Descriptor: | 1,2-ETHANEDIOL, Proline--tRNA ligase, [(2~{R},3~{S})-2-[3-(7-bromanyl-6-chloranyl-4-oxidanylidene-quinazolin-3-yl)-2-oxidanylidene-propyl]piperidin-3-yl] ~{N}-[4-[[3-(2,3-dihydro-1~{H}-inden-2-ylcarbamoyl)pyrazin-2-yl]carbamoyl]piperazin-1-yl]sulfonylcarbamate | | Authors: | Johansson, C, Tye, M, Payne, N.C, Mazitschek, R, Oppermann, U.C.T. | | Deposit date: | 2021-11-22 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of Prolyl-tRNA synthetase (ProRS, Proline-tRNA ligase) from Plasmodium falciparum in complex with MAT436
To Be Published
|
|
6Q02
 
 | |
6JGS
 
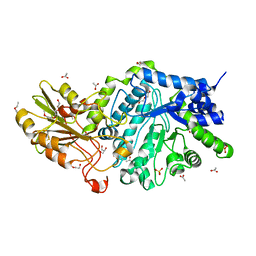 | | Crystal structure of barley exohydrolaseI W434Y mutant in complex with 4I,4III,4V-S-trithiocellohexaose. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, BETA-D-GLUCAN GLUCOHYDROLASE ISOENZYME EXO1, ... | | Authors: | Luang, S, Streltsov, V.A, Hrmova, M. | | Deposit date: | 2019-02-14 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The evolutionary advantage of an aromatic clamp in plant family 3 glycoside exo-hydrolases.
Nat Commun, 13, 2022
|
|
6FC4
 
 | |
5UPF
 
 | |
