9IHQ
 
 | |
9IHO
 
 | |
4K6C
 
 | |
5TEI
 
 | | Structure of human ALDH1A1 with inhibitor CM039 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-5-(2-methylphenyl)-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ALDH1A1 with inhibitor CM039
To Be Published
|
|
4CCJ
 
 | | 60S ribosomal protein L8 histidine hydroxylase (NO66) in apo form | | Descriptor: | 1,2-ETHANEDIOL, BIFUNCTIONAL LYSINE-SPECIFIC DEMETHYLASE AND HISTIDYL-HYDROXYLASE NO66 | | Authors: | Chowdhury, R, Ge, W, Clifton, I.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
3UV3
 
 | | Ec_IspH in complex with but-2-ynyl diphosphate (1086) | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER, but-2-yn-1-yl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-29 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
7CB7
 
 | | 1.7A resolution structure of SARS-CoV-2 main protease (Mpro) in complex with broad-spectrum coronavirus protease inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Wang, Y.C, Yang, C.S, Hou, M.H, Tsai, C.L, Chou, Y.Z, Chen, Y, Hung, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug.
Am J Cancer Res, 10, 2020
|
|
4CCQ
 
 | | Crystal structure of the thioredoxin reductase from Entamoeba histolytica with NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Parsonage, D, Kells, P.M, Hirata, K, Debnath, A, Poole, L.B, McKerrow, J.H, Reed, S.L, Podust, L.M. | | Deposit date: | 2013-10-24 | | Release date: | 2014-11-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-Ray Structures of Thioredoxin and Thioredoxin Reductase from Entamoeba Histolytica and Prevailing Hypothesis of the Mechanism of Auranofin Action.
J.Struct.Biol., 194, 2016
|
|
6RFR
 
 | | Cryo-EM structure of respiratory complex I from Yarrowia lipolytica at 3.2 A resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Acyl carrier protein ACPM1 of NADH:Ubiquinone Oxidoreductase (Complex I), ... | | Authors: | Parey, K, Vonck, J. | | Deposit date: | 2019-04-16 | | Release date: | 2019-12-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of respiratory complex I: Mechanism, assembly, and disease.
Sci Adv, 5, 2019
|
|
6F9J
 
 | | Crystal structure of Barley Beta-Amylase complexed with 4-O-alpha-D-mannopyranosyl-(1-deoxynojirimycin) | | Descriptor: | Beta-amylase, CHLORIDE ION, alpha-D-mannopyranose-(1-4)-1-DEOXYNOJIRIMYCIN | | Authors: | Moncayo, M.A, Rodrigues, L.L, Stevenson, C.E.M, Ruzanski, C, Rejzek, M, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2017-12-14 | | Release date: | 2019-01-30 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Synthesis, biological and structural analysis of prospective glycosyl-iminosugar prodrugs: impact on germination
To be published
|
|
6B5G
 
 | |
3UTC
 
 | | Ec_IspH in complex with (E)-4-hydroxybut-3-enyl diphosphate | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Li, K, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-11-25 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3UW3
 
 | |
9GRX
 
 | | NDH-PSI-LHCI supercomplex from S. oleracea | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Introini, B, Hahn, A, Kuehlbrandt, W. | | Deposit date: | 2024-09-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Cryo-EM structure of the NDH-PSI-LHCI supercomplex from Spinacia oleracea.
Nat.Struct.Mol.Biol., 32, 2025
|
|
6BAW
 
 | | Structure of GRP94 with a selective resorcinylic inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, Endoplasmin, PHOSPHATE ION, ... | | Authors: | Que, N.L.S, Gewirth, D.T. | | Deposit date: | 2017-10-16 | | Release date: | 2018-04-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
J. Med. Chem., 61, 2018
|
|
6HU9
 
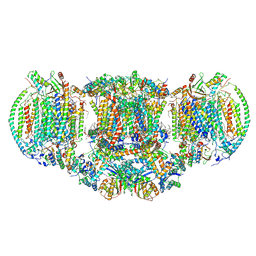 | | III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Hartley, A.M, Pinotsis, N, Marechal, A. | | Deposit date: | 2018-10-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat. Struct. Mol. Biol., 26, 2019
|
|
7JZ2
 
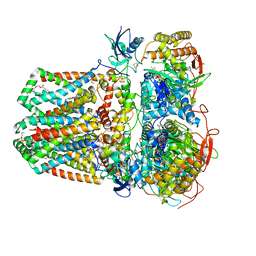 | | Succinate: quinone oxidoreductase SQR from E.coli K12 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lyu, M, Su, C.-C, Morgan, C.E, Bolla, J.R, Robinson, C.V, Yu, E.W. | | Deposit date: | 2020-09-01 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A 'Build and Retrieve' methodology to simultaneously solve cryo-EM structures of membrane proteins.
Nat.Methods, 18, 2021
|
|
9I9L
 
 | | Structure of Far-Red Photosystem I from C. thermalis PCC 7203 | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Consoli, G, Tufaill, F, Murray, J.W, Fantuzzi, A, Rutherford, A.W. | | Deposit date: | 2025-02-06 | | Release date: | 2025-10-22 | | Method: | ELECTRON MICROSCOPY (1.89 Å) | | Cite: | Locating the missing chlorophylls f in far-red photosystem I.
Science, 2025
|
|
6DK1
 
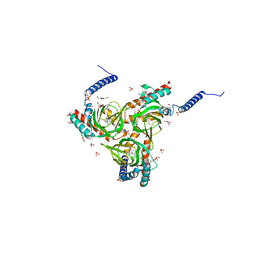 | | Human sigma-1 receptor bound to (+)-pentazocine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S,6S,11S)-6,11-dimethyl-3-(3-methylbut-2-en-1-yl)-1,2,3,4,5,6-hexahydro-2,6-methano-3-benzazocin-8-ol, GLYCEROL, ... | | Authors: | Schmidt, H.R, Kruse, A.C. | | Deposit date: | 2018-05-28 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Structural basis for sigma1receptor ligand recognition.
Nat. Struct. Mol. Biol., 25, 2018
|
|
3UWM
 
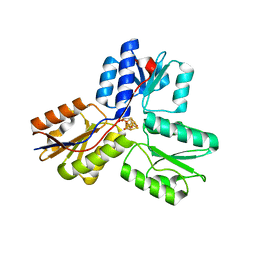 | | Ec_IspH in complex with 4-oxobutyl diphosphate (1302) | | Descriptor: | (3E)-4-hydroxybut-3-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, 4-oxobutyl trihydrogen diphosphate, ... | | Authors: | Span, I, Wang, K, Wang, W, Zhang, Y, Bacher, A, Eisenreich, W, Schulz, C, Oldfield, E, Groll, M. | | Deposit date: | 2011-12-02 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of acetylene hydratase activity of the iron-sulphur protein IspH.
Nat Commun, 3, 2012
|
|
3AT3
 
 | | Crystal structure of CK2alpha with pyradine derivative | | Descriptor: | (1-{6-[6-(cyclopentylamino)-1H-indazol-1-yl]pyrazin-2-yl}-1H-pyrrol-3-yl)acetic acid, Casein kinase II subunit alpha | | Authors: | Kinoshita, T. | | Deposit date: | 2010-12-23 | | Release date: | 2011-11-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A detailed thermodynamic profile of cyclopentyl and isopropyl derivatives binding to CK2 kinase
Mol.Cell.Biochem., 356, 2011
|
|
5O2B
 
 | | Crystal structure of WNK3 kinase domain in a diphosphorylated state and in a complex with the inhibitor PP-121 | | Descriptor: | 1-cyclopentyl-3-(1H-pyrrolo[2,3-b]pyridin-5-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, SODIUM ION, Serine/threonine-protein kinase WNK3 | | Authors: | Pinkas, D.M, Bufton, J.C, Newman, J.A, Borkowska, O, Chalk, R, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-05-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.038 Å) | | Cite: | Crystal structure of WNK3 kinase domain in a diphosphorylated state and in a complex with the inhibitor PP-121
To Be Published
|
|
4IIU
 
 | | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Osinski, T, Zheng, H, Shumilin, I, Shabalin, I, Shatsman, S, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-12-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a putative 3-oxoacyl-[acyl-carrier protein]reductase from Escherichia coli strain CFT073 complexed with NADP+ at 2.1 A resolution
To be Published
|
|
7KI8
 
 | |
7KI9
 
 | |
