3PDX
 
 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
5WEH
 
 | | Cytochrome c oxidase from Rhodobacter sphaeroides in the reduced state | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Aa3-type cytochrome c oxidase subunit IV, CALCIUM ION, ... | | Authors: | Liu, J, Ferguson-Miller, F, Ling, Q, Hiser, C. | | Deposit date: | 2017-07-10 | | Release date: | 2017-09-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Role of conformational change and K-path ligands in controlling cytochrome c oxidase activity.
Biochem. Soc. Trans., 45, 2017
|
|
4P2X
 
 | | Swapped Dimer of Mycobacterial Adenylyl cyclase Rv1625c: Form 2 | | Descriptor: | Adenylate cyclase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Barathy, D.V, Mattoo, R, Visweswariah, S.S, Suguna, K. | | Deposit date: | 2014-03-05 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | New structural forms of a mycobacterial adenylyl cyclase Rv1625c.
Iucrj, 1, 2014
|
|
4ARV
 
 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
5DUH
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Sulfoxide-bridged Oxabicyclic Heptene Sulfonate (SOBHS)-3 analog phenyl (1S,2S,4S,7S)-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate 7-oxide | | Descriptor: | Estrogen receptor, Nuclear receptor coactivator 2, phenyl (1S,2S,4S,7S)-5,6-bis(4-hydroxy-3-methylphenyl)-7-thiabicyclo[2.2.1]hept-5-ene-2-sulfonate 7-oxide | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-18 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DXM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol | | Descriptor: | 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5YFM
 
 | |
5TPI
 
 | | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator (LysR family), SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.
To Be Published
|
|
5Y5I
 
 | | Time-resolved SFX structure of cytochrome P450nor: 20 ms after photo-irradiation of caged NO in the presence of NADH (NO-bound state), light data | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5WHL
 
 | |
7Y00
 
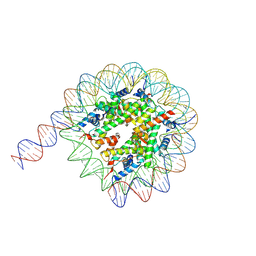 | | Cryo-EM structure of the nucleosome containing 169 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (169-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZY
 
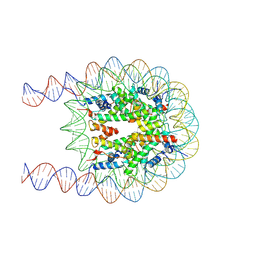 | | Cryo-EM structure of the nucleosome containing 193 base-pair DNA with a p53 target sequence | | Descriptor: | DNA (193-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
3URH
 
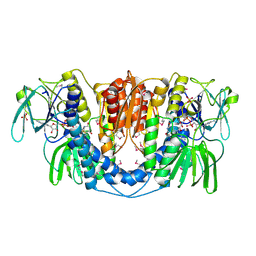 | | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021 | | Descriptor: | 1,2-ETHANEDIOL, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kumaran, D, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-11-22 | | Release date: | 2011-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a dihydrolipoamide dehydrogenase from Sinorhizobium meliloti 1021
TO BE PUBLISHED
|
|
7XZZ
 
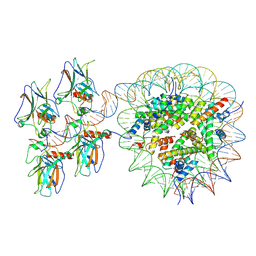 | | Cryo-EM structure of the nucleosome in complex with p53 | | Descriptor: | Cellular tumor antigen p53, DNA (169-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
7XZX
 
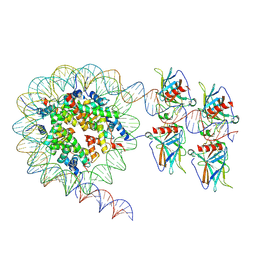 | | Cryo-EM structure of the nucleosome in complex with p53 DNA-binding domain | | Descriptor: | Cellular tumor antigen p53, DNA (193-MER), Histone H2A type 1-B/E, ... | | Authors: | Nishimura, M, Nozawa, K, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.53 Å) | | Cite: | Structural basis for p53 binding to its nucleosomal target DNA sequence.
Pnas Nexus, 1, 2022
|
|
2W9H
 
 | |
3ITJ
 
 | | Crystal structure of Saccharomyces cerevisiae thioredoxin reductase 1 (Trr1) | | Descriptor: | CITRIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Thioredoxin reductase 1 | | Authors: | Oliveira, M.A, Discola, K.F, Alves, S.V, Medrano, F.J, Guimaraes, B.G, Netto, L.E.S. | | Deposit date: | 2009-08-28 | | Release date: | 2010-03-31 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the specificity of thioredoxin reductase-thioredoxin interactions. A structural and functional investigation of the yeast thioredoxin system.
Biochemistry, 49, 2010
|
|
1A7A
 
 | | STRUCTURE OF HUMAN PLACENTAL S-ADENOSYLHOMOCYSTEINE HYDROLASE: DETERMINATION OF A 30 SELENIUM ATOM SUBSTRUCTURE FROM DATA AT A SINGLE WAVELENGTH | | Descriptor: | (1'R,2'S)-9-(2-HYDROXY-3'-KETO-CYCLOPENTEN-1-YL)ADENINE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, S-ADENOSYLHOMOCYSTEINE HYDROLASE | | Authors: | Turner, M.A, Yuan, C.-S, Borchardt, R.T, Hershfield, M.S, Smith, G.D, Howell, P.L. | | Deposit date: | 1998-03-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure determination of selenomethionyl S-adenosylhomocysteine hydrolase using data at a single wavelength.
Nat.Struct.Biol., 5, 1998
|
|
8J5K
 
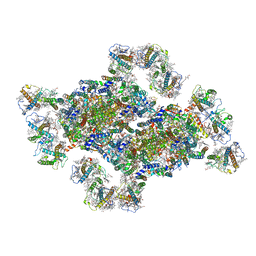 | | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15-octaen-17-ynyl]cyclohex-3-en-1-ol, (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, (3S,3'S,5R,5'R,6S,6'R,8'R)-3,5'-dihydroxy-8-oxo-6',7'-didehydro-5,5',6,6',7,8-hexahydro-5,6-epoxy-beta,beta-caroten-3'- yl acetate, ... | | Authors: | Shen, L.L, Li, Z.H, Shen, J.R, Wang, W.D. | | Deposit date: | 2023-04-23 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural insights into photosystem II supercomplex and trimeric FCP antennae of a centric diatom Cyclotella meneghiniana.
Nat Commun, 14, 2023
|
|
5F61
 
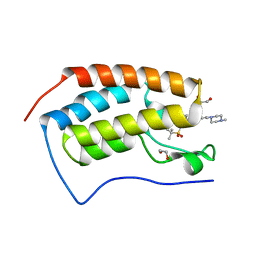 | | Crystal structure of the first bromodomain of human BRD4 in complex with MA4-022-1 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ~{N}-[3-[[2-[[3-fluoranyl-4-(4-methylpiperazin-1-yl)phenyl]amino]-5-methyl-pyrimidin-4-yl]amino]phenyl]-2-methyl-propane-2-sulfonamide | | Authors: | Ember, S.W, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2015-12-04 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Potent Dual BET Bromodomain-Kinase Inhibitors as Value-Added Multitargeted Chemical Probes and Cancer Therapeutics.
Mol. Cancer Ther., 16, 2017
|
|
5Y5J
 
 | | Time-resolved SFX structure of cytochrome P450nor: dark-2 data in the presence of NADH (resting state) | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tosha, T, Nomura, T, Nishida, T, Saeki, N, Okubayashi, K, Yamagiwa, R, Sugahara, M, Nakane, T, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Mizohata, E, Yamashita, A, Kanematsu, Y, Takano, Y, Nango, E, Tanaka, R, Nureki, O, Ikemoto, Y, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Iwata, S, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6LY4
 
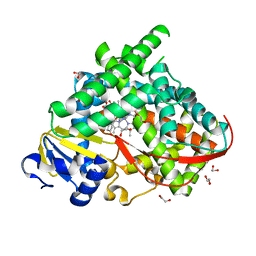 | | The crystal structure of the BM3 mutant LG-23 in complex with testosterone | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, IMIDAZOLE, ... | | Authors: | Peng, Y, Chen, J, Zhou, J, Li, A, ReetZ, M.T. | | Deposit date: | 2020-02-13 | | Release date: | 2020-04-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Regio- and Stereoselective Steroid Hydroxylation at C7 by Cytochrome P450 Monooxygenase Mutants.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3INV
 
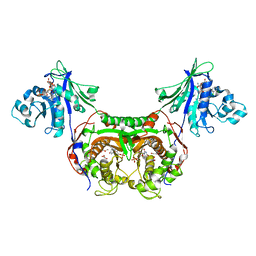 | | Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase COMPLEXED WITH NADPH, dUMP AND C-448 ANTIFOLATE | | Descriptor: | 1-[3-(2,3-dichlorophenoxy)propoxy]-6,6-dimethyl-1,6-dihydro-1,3,5-triazine-2,4-diamine, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase, ... | | Authors: | Chitnumsub, P, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2009-08-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural basis of antifolate inhibition of Trypanosoma cruzi Dihydrofolate Reductase-Thymidylate Synthase
To be Published
|
|
6M5X
 
 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
7B74
 
 | | Chimeric Streptavidin With A Dimerization Domain For Artificial Transfer Hydrogenation | | Descriptor: | Streptavidin,Superoxide dismutase [Cu-Zn],Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2020-12-09 | | Release date: | 2021-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Spiers Memorial Lecture: Shielding the active site: a streptavidin superoxide-dismutase chimera as a host protein for asymmetric transfer hydrogenation.
Faraday Disc.Chem.Soc, 2023
|
|
