6Y2L
 
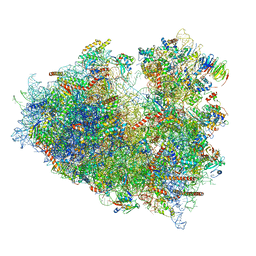 | | Structure of human ribosome in POST state | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Bhaskar, V, Schenk, A.D, Cavadini, S, von Loeffelholz, O, Natchiar, S.K, Klaholz, B.P, Chao, J.A. | | Deposit date: | 2020-02-16 | | Release date: | 2020-04-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Dynamics of uS19 C-Terminal Tail during the Translation Elongation Cycle in Human Ribosomes.
Cell Rep, 31, 2020
|
|
8FQN
 
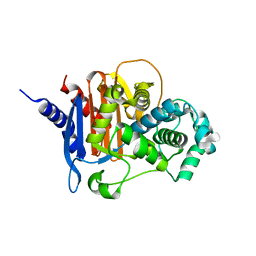 | | apo ADC-33 beta-lactamase | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION | | Authors: | Powers, R.A, Wallar, B.J, June, C.M, Fish, E.R. | | Deposit date: | 2023-01-06 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.256 Å) | | Cite: | Synthesis of a Novel Boronic Acid Transition State Inhibitor, MB076: A Heterocyclic Triazole Effectively Inhibits Acinetobacter -Derived Cephalosporinase Variants with an Expanded-Substrate Spectrum.
J.Med.Chem., 66, 2023
|
|
3FH7
 
 | |
7D5C
 
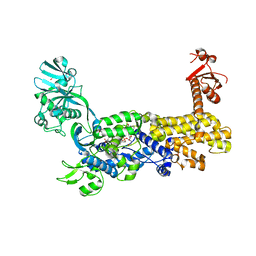 | | IleRS in complex with a tRNA site inhibitor | | Descriptor: | (2E,4S,5S,6E,8E)-10-[(2S,3R,6S,8R,9S)-3-butyl-9-methyl-2-[(1E,3E)-3-methyl-5-oxidanyl-5-oxidanylidene-penta-1,3-dienyl]-3-(4-oxidanyl-4-oxidanylidene-butanoyl)oxy-1,7-dioxaspiro[5.5]undecan-8-yl]-4,8-dimethyl-5-oxidanyl-deca-2,6,8-trienoic acid, 1,2-ETHANEDIOL, Isoleucine--tRNA ligase, ... | | Authors: | Chen, B, Luo, S, Zhou, H. | | Deposit date: | 2020-09-25 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Inhibitory mechanism of reveromycin A at the tRNA binding site of a class I synthetase.
Nat Commun, 12, 2021
|
|
5FOY
 
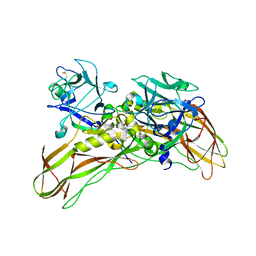 | | De novo structure of the binary mosquito larvicide BinAB at pH 7 | | Descriptor: | 41.9 KDA INSECTICIDAL TOXIN, LARVICIDAL TOXIN 51 KDA PROTEIN | | Authors: | Colletier, J.P, Sawaya, M.R, Gingery, M, Rodriguez, J.A, Cascio, D, Brewster, A.S, Michels-Clark, T, Boutet, S, Williams, G.J, Messerschmidt, M, DePonte, D.P, Sierra, R.G, Laksmono, H, Koglin, J.E, Hunter, M.S, W Park, H, Uervirojnangkoorn, M, Bideshi, D.L, Brunger, A.T, Federici, B.A, Sauter, N.K, Eisenberg, D.S. | | Deposit date: | 2015-11-26 | | Release date: | 2016-10-05 | | Last modified: | 2019-08-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | De Novo Phasing with X-Ray Laser Reveals Mosquito Larvicide Binab Structure.
Nature, 539, 2016
|
|
3EYE
 
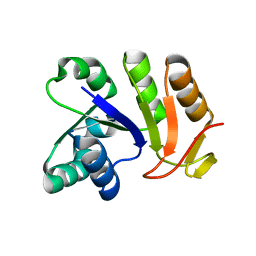 | | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli | | Descriptor: | PTS system N-acetylgalactosamine-specific IIB component 1 | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Do, J, Sampathkumar, P, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-10-20 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of PTS system N-acetylgalactosamine-specific IIB component 1 from Escherichia coli
To be Published
|
|
8FIL
 
 | | Zinc-free APOBEC3A (inactive E72A mutant) in complex with TTC-hairpin DNA substrate | | Descriptor: | CHLORIDE ION, DNA (5'-D(*TP*GP*CP*GP*CP*TP*TP*CP*GP*CP*GP*CP*T)-3'), DNA dC->dU-editing enzyme APOBEC-3A, ... | | Authors: | Harjes, S, Jameson, G.B, Harjes, E, Filichev, V.V, Kurup, H.M. | | Deposit date: | 2022-12-16 | | Release date: | 2023-09-06 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure-guided inhibition of the cancer DNA-mutating enzyme APOBEC3A.
Nat Commun, 14, 2023
|
|
2O8H
 
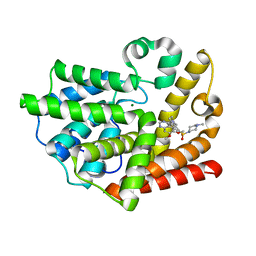 | | Crystal structure of the catalytic domain of rat phosphodiesterase 10A | | Descriptor: | 6,7-DIMETHOXY-4-{8-[(4-METHYLPIPERAZIN-1-YL)SULFONYL]-3,4-DIHYDROISOQUINOLIN-2(1H)-YL}QUINAZOLINE, MAGNESIUM ION, Phosphodiesterase-10A, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-01-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors
J.Med.Chem., 50, 2007
|
|
1TV4
 
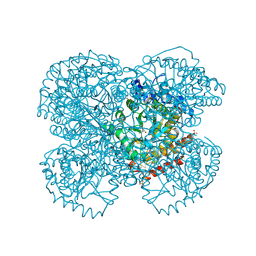 | | Crystal structure of the sulfite MtmB complex | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, Monomethylamine methyltransferase mtmB1, SULFATE ION | | Authors: | Hao, B, Zhao, G, Kang, P.T, Soares, J.A, Ferguson, T.K, Gallucci, J, Krzycki, J.A, Chan, M.K. | | Deposit date: | 2004-06-26 | | Release date: | 2004-10-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reactivity and chemical synthesis of L-pyrrolysine- the 22(nd) genetically encoded amino acid
Chem.Biol., 11, 2004
|
|
6T6W
 
 | |
3EW7
 
 | | Crystal structure of the Lmo0794 protein from Listeria monocytogenes. Northeast Structural Genomics Consortium target LmR162. | | Descriptor: | Lmo0794 protein | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Wang, D, Foote, E.L, Ciccosanti, C, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of the Lmo0794 protein from Listeria monocytogenes.
To be Published
|
|
3EWD
 
 | | Crystal structure of adenosine deaminase mutant (delta Asp172) from Plasmodium vivax in complex with MT-coformycin | | Descriptor: | (8R)-3-(5-S-methyl-5-thio-beta-D-ribofuranosyl)-3,6,7,8-tetrahydroimidazo[4,5-d][1,3]diazepin-8-ol, Adenosine deaminase, ZINC ION | | Authors: | Schramm, V.L, Almo, S.C, Cassera, M.B, Ho, M.C. | | Deposit date: | 2008-10-14 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and metabolic specificity of methylthiocoformycin for malarial adenosine deaminases.
Biochemistry, 48, 2009
|
|
3F68
 
 | | Thrombin Inhibition | | Descriptor: | Hirudin variant-2, N-acetyl-3-cyclohexyl-D-alanyl-N-(3-chlorobenzyl)-L-prolinamide, Prothrombin, ... | | Authors: | Baum, B, Heine, A, Klebe, G. | | Deposit date: | 2008-11-05 | | Release date: | 2009-10-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Think twice: understanding the high potency of bis(phenyl)methane inhibitors of thrombin.
J.Mol.Biol., 391, 2009
|
|
1TX9
 
 | | gpd prior to capsid assembly | | Descriptor: | Scaffolding protein D | | Authors: | Morais, M.C, Fisher, M, Kanamaru, K, Fane, B.A, Rossmann, M.G. | | Deposit date: | 2004-06-24 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Conformational switching by the scaffolding protein D directs the assembly of bacteriophage phiX174
Mol.Cell, 15, 2004
|
|
3EXN
 
 | | Crystal structure of acetyltransferase from Thermus thermophilus HB8 | | Descriptor: | ACETYL COENZYME *A, CHLORIDE ION, Probable acetyltransferase | | Authors: | Nocek, B, Hatzos, C, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-16 | | Release date: | 2009-01-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of acetyltransferase from Thermus thermophilus HB8
To be Published
|
|
8FLJ
 
 | | Cas1-Cas2/3 integrase and IHF bound to CRISPR leader, repeat and foreign DNA | | Descriptor: | CRISPR leader and repeat, anti-sense strand of DNA, CRISPR leader, ... | | Authors: | Santiago-Frangos, A, Henriques, W.S, Wiegand, T, Gauvin, C, Buyukyoruk, M, Neselu, K, Eng, E.T, Lander, G.C, Wiedenheft, B. | | Deposit date: | 2022-12-21 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure reveals why genome folding is necessary for site-specific integration of foreign DNA into CRISPR arrays.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2O4X
 
 | | Crystal structure of human P100 tudor domain | | Descriptor: | Staphylococcal nuclease domain-containing protein 1 | | Authors: | Shaw, N, Zhao, M, Cheng, C, Xu, H, Yang, J, Silvennoinen, O, Rao, Z, Wang, B.C, Liu, Z.J. | | Deposit date: | 2006-12-05 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human P100 tudor domain
To be Published
|
|
3FDL
 
 | | Bim BH3 peptide in complex with Bcl-xL | | Descriptor: | Apoptosis regulator Bcl-X, Bcl-2-like protein 11 | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2008-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | High-Resolution Structural Characterization of a Helical alpha/beta-Peptide Foldamer Bound to the Anti-Apoptotic Protein Bcl-x(L)
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
5AJX
 
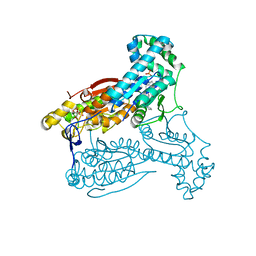 | | Human PFKFB3 in complex with an indole inhibitor 3 | | Descriptor: | (2S)-N-[4-[3-cyano-1-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]indol-5-yl]oxyphenyl]pyrrolidine-2-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
2O63
 
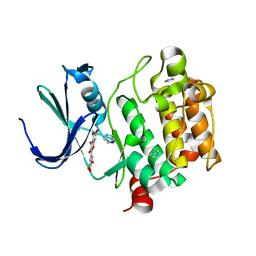 | | Crystal structure of Pim1 with Myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, IMIDAZOLE, Proto-oncogene serine/threonine-protein kinase Pim-1 | | Authors: | Holder, S, Zemskova, M, Zhang, C, Tabrizizad, M, Bremer, R, Neidigh, J.W, Lilly, M.B. | | Deposit date: | 2006-12-06 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase.
Mol.Cancer Ther., 6, 2007
|
|
3EZR
 
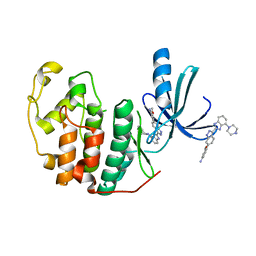 | | CDK-2 with indazole inhibitor 17 bound at its active site | | Descriptor: | 3-methoxy-4-{3-[4-(4-methylpiperazin-1-yl)-1H-benzimidazol-2-yl]-1H-indazol-6-yl}aniline, Cell division protein kinase 2 | | Authors: | Kiefer, J.R, Day, J.E, Caspers, N.L, Mathis, K.J, Kretzmer, K.K, Weinberg, R.A, Reitz, B.A, Stegeman, R.A, Trujillo, J.I, Huang, W, Thorarensen, A, Xing, L, Wrightstone, A, Christine, L, Compton, R, Li, X. | | Deposit date: | 2008-10-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 2-(6-Phenyl-1H-indazol-3-yl)-1H-benzo[d]imidazoles: Design and synthesis of a potent and isoform selective PKC-zeta inhibitor
Bioorg.Med.Chem.Lett., 19, 2009
|
|
6W2H
 
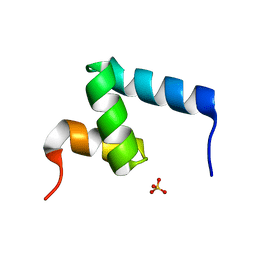 | | Crystal Structure of the Internal UBA Domain of HHR23A | | Descriptor: | SULFATE ION, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Residual Structure in the Denatured State of the Fast-Folding UBA(1) Domain from the Human DNA Excision Repair Protein HHR23A.
Biochemistry, 61, 2022
|
|
2O6Q
 
 | |
4J6A
 
 | | Crystal structure of Ribonuclease A soaked in 40% 2,2,2-Trifluoroethanol: One of twelve in MSCS set | | Descriptor: | Ribonuclease pancreatic, SULFATE ION, TRIFLUOROETHANOL | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
1U2P
 
 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Protein Tyrosine Phosphatase (MPtpA) at 1.9A resolution | | Descriptor: | CHLORIDE ION, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
