1R7R
 
 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
1NFB
 
 | |
1OL5
 
 | | Structure of Aurora-A 122-403, phosphorylated on Thr287, Thr288 and bound to TPX2 1-43 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RESTRICTED EXPRESSION PROLIFERATION ASSOCIATED PROTEIN 100, ... | | Authors: | Bayliss, R, Conti, E. | | Deposit date: | 2003-08-06 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Aurora-A Activation by Tpx2 at the Mitotic Spindle
Mol.Cell, 12, 2003
|
|
1OL7
 
 | |
1Q60
 
 | | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA | | Descriptor: | General transcription factor II-I | | Authors: | Doi-Katayama, Y, Hayashi, F, Hirota, H, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-12 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of RSGI RUH-004, a GTF2I domain in Mouse cDNA
To be published
|
|
1PQ9
 
 | | HUMAN LXR BETA HORMONE RECEPTOR COMPLEXED WITH T0901317 COMPLEX | | Descriptor: | 1,1,1,3,3,3-HEXAFLUORO-2-{4-[(2,2,2-TRIFLUOROETHYL)AMINO]PHENYL}PROPAN-2-OL, Oxysterols receptor LXR-beta, benzenesulfonic acid | | Authors: | Farnegardh, M, Bonn, T, Sun, S, Ljunggren, J, Ahola, H, Wilhelmsson, A, Gustafsson, J.-A, Carlquist, M. | | Deposit date: | 2003-06-18 | | Release date: | 2003-09-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The three-dimensional structure of the liver X receptor beta reveals a flexible ligand-binding pocket that can accommodate fundamentally different ligands.
J.Biol.Chem., 278, 2003
|
|
1Q02
 
 | | NMR structure of the UBA domain of p62 (SQSTM1) | | Descriptor: | sequestosome 1 | | Authors: | Ciani, B, Layfield, R, Cavey, J.R, Sheppard, P.W, Searle, M.S. | | Deposit date: | 2003-07-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ubiquitin-associated Domain of p62 (SQSTM1) and Implications for Mutations That Cause Paget's
Disease of Bone
J.Biol.Chem., 278, 2003
|
|
1RHO
 
 | | STRUCTURE OF RHO GUANINE NUCLEOTIDE DISSOCIATION INHIBITOR | | Descriptor: | RHO GDP-DISSOCIATION INHIBITOR 1, SULFATE ION | | Authors: | Keep, N.H, Moody, P.C.E, Roberts, G.C.K. | | Deposit date: | 1996-10-12 | | Release date: | 1997-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A modulator of rho family G proteins, rhoGDI, binds these G proteins via an immunoglobulin-like domain and a flexible N-terminal arm.
Structure, 5, 1997
|
|
1QG2
 
 | | CANINE GDP-RAN R76E MUTANT | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (RAN) | | Authors: | Kent, H.M, Moore, M.S, Quimby, B.B, Baker, A.M.E, McCoy, A.J, Murphy, G.A, Corbett, A.H, Stewart, M. | | Deposit date: | 1999-04-20 | | Release date: | 1999-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Engineered mutants in the switch II loop of Ran define the contribution made by key residues to the interaction with nuclear transport factor 2 (NTF2) and the role of this interaction in nuclear protein import.
J.Mol.Biol., 289, 1999
|
|
1RRP
 
 | | STRUCTURE OF THE RAN-GPPNHP-RANBD1 COMPLEX | | Descriptor: | MAGNESIUM ION, NUCLEAR PORE COMPLEX PROTEIN NUP358, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Vetter, I.R, Nowak, C, Nishimoto, T, Kuhlmann, J, Wittinghofer, A. | | Deposit date: | 1999-01-15 | | Release date: | 1999-05-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport.
Nature, 398, 1999
|
|
1SAV
 
 | | HUMAN ANNEXIN V WITH PROLINE SUBSTITUTION BY THIOPROLINE | | Descriptor: | ANNEXIN V, CALCIUM ION | | Authors: | Medrano, F.J, Minks, C, Budisa, N, Huber, R. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal and molecular structure of human annexin V after refinement. Implications for structure, membrane binding and ion channel formation of the annexin family of proteins.
J.Mol.Biol., 223, 1992
|
|
3MB4
 
 | | Crystal Structure of the fifth Bromodomain of Human Poly-bromodomain containing protein 1 (PB1) with NMP | | Descriptor: | 1,2-ETHANEDIOL, 1-methylpyrrolidin-2-one, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Keates, T, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-25 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3N5E
 
 | | Crystal Structure of human thymidylate synthase bound to a peptide inhibitor | | Descriptor: | SULFATE ION, Synthetic peptide LR, Thymidylate synthase | | Authors: | Pozzi, C, Cardinale, D, Guaitoli, G, Tondi, D, Luciani, R, Myllykallio, H, Ferrari, S, Costi, M.P, Mangani, S. | | Deposit date: | 2010-05-25 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3NQJ
 
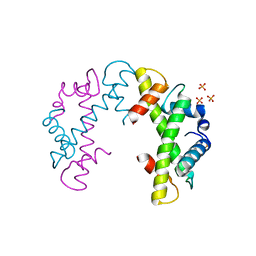 | | Crystal structure of (CENP-A/H4)2 heterotetramer | | Descriptor: | Histone H3-like centromeric protein A, Histone H4, PHOSPHATE ION | | Authors: | Sekulic, N, Black, B.E. | | Deposit date: | 2010-06-29 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres.
Nature, 467, 2010
|
|
1JUN
 
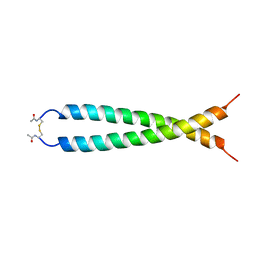 | |
1JUJ
 
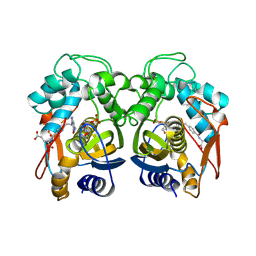 | | Human Thymidylate Synthase Bound to dUMP and LY231514, a Pyrrolo(2,3-d)pyrimidine-based Antifolate | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-{4-[2-(2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDIN-5-YL)-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Sayre, P.H, Finer-Moore, J.S, Fritz, T.A, Biermann, D, Gates, S.B, MacKellar, W.C, Patel, V.F, Stroud, R.M. | | Deposit date: | 2001-08-24 | | Release date: | 2001-09-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases.
J.Mol.Biol., 313, 2001
|
|
1KBQ
 
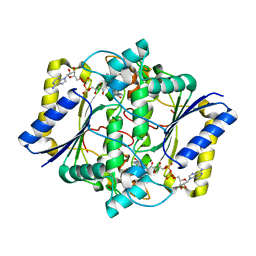 | | Complex of Human NAD(P)H quinone Oxidoreductase with 5-methoxy-1,2-dimethyl-3-(4-nitrophenoxymethyl)indole-4,7-dione (ES936) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(4-NITROPHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
3O2T
 
 | |
1KBO
 
 | | Complex of Human recombinant NAD(P)H:Quinone Oxide reductase type 1 with 5-methoxy-1,2-dimethyl-3-(phenoxymethyl)indole-4,7-dione (ES1340) | | Descriptor: | 5-METHOXY-1,2-DIMETHYL-3-(PHENOXYMETHYL)INDOLE-4,7-DIONE, FLAVIN-ADENINE DINUCLEOTIDE, NAD(P)H dehydrogenase [quinone] 1 | | Authors: | Faig, M, Bianchet, M.A, Amzel, L.M. | | Deposit date: | 2001-11-06 | | Release date: | 2002-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Characterization of a mechanism-based inhibitor of NAD(P)H:quinone oxidoreductase 1 by biochemical, X-ray crystallographic, and mass spectrometric approaches.
Biochemistry, 40, 2001
|
|
3NNC
 
 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*UP*GP*UP*GP*UP*GP*UP*UP*GP*UP*GP*UP*G)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2005 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
3O50
 
 | | Crystal structure of benzamide 9 bound to AuroraA | | Descriptor: | N-{3-methyl-4-[(3-pyrimidin-4-ylpyridin-2-yl)oxy]phenyl}-3-(trifluoromethyl)benzamide, cDNA FLJ58295, highly similar to Serine/threonine-protein kinase 6 | | Authors: | Huang, X. | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a potent, selective, and orally bioavailable pyridinyl-pyrimidine phthalazine aurora kinase inhibitor.
J.Med.Chem., 53, 2010
|
|
1KDX
 
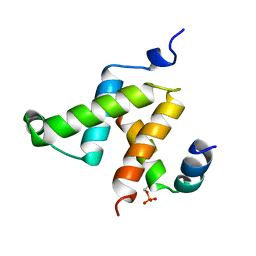 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
3NRM
 
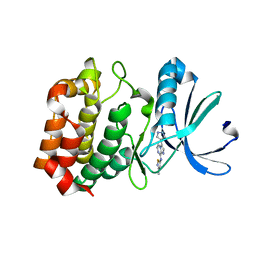 | | Imidazo[1,2-a]pyrazine-based Aurora Kinase Inhibitors | | Descriptor: | N-(3-methylisothiazol-5-yl)-3-(1H-pyrazol-4-yl)imidazo[1,2-a]pyrazin-8-amine, Serine/threonine-protein kinase 6 | | Authors: | Hruza, A. | | Deposit date: | 2010-06-30 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery of imidazo[1,2-a]pyrazine-based Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3NNA
 
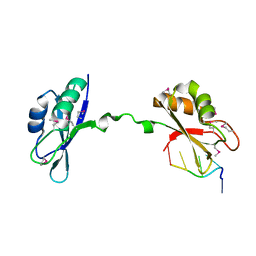 | | Crystal Structure of CUGBP1 RRM1/2-RNA Complex | | Descriptor: | CUGBP Elav-like family member 1, RNA (5'-R(*GP*UP*UP*GP*UP*UP*UP*UP*GP*UP*UP*U)-3') | | Authors: | Teplova, M, Song, J, Gaw, H, Teplov, A, Patel, D.J. | | Deposit date: | 2010-06-23 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural Insights into RNA Recognition by the Alternate-Splicing Regulator CUG-Binding Protein 1.
Structure, 18, 2010
|
|
1M9F
 
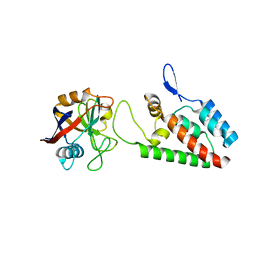 | | X-ray crystal structure of Cyclophilin A/HIV-1 CA N-terminal domain (1-146) M-type H87A,A88M Complex. | | Descriptor: | Cyclophilin A, HIV-1 Capsid | | Authors: | Howard, B.R, Vajdos, F.F, Li, S, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2002-07-28 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural insights into the catalytic mechanism of cyclophilin A
Nat.Struct.Biol., 10, 2003
|
|
