8EM5
 
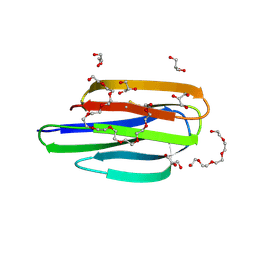 | | Mycobacterium thermoresistible MmpS5 | | Descriptor: | DODECAETHYLENE GLYCOL, GLYCEROL, IODIDE ION, ... | | Authors: | Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-09-26 | | Release date: | 2023-09-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The structure of Mycobacterium thermoresistibile MmpS5 reveals a conserved disulfide bond across mycobacteria.
Metallomics, 16, 2024
|
|
8P6G
 
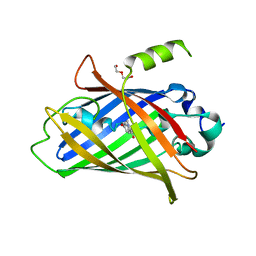 | | Crystal structure of the improved version of the Genetically Encoded Green Calcium Indicator YTnC2-5 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Genetically Encoded Green Calcium Indicator YTnC2-5 based on Troponin C from toadfish | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Subach, O.M, Vlaskina, A.V, Agapova, Y.K, Popov, V.O, Subach, F.V. | | Deposit date: | 2023-05-26 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | YTnC2, an improved genetically encoded green calcium indicator based on toadfish troponin C.
Febs Open Bio, 13, 2023
|
|
8P37
 
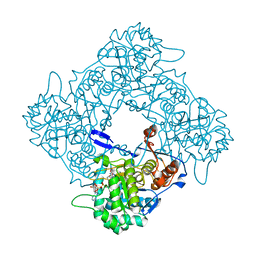 | | Structure a catalytically inactive mutant of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
7UVA
 
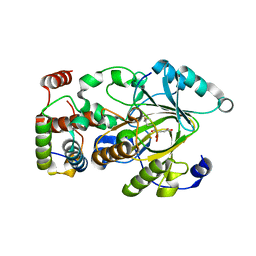 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
6X7E
 
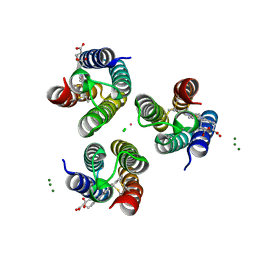 | | Co-bound structure of an engineered protein trimer, TriCyt3, with delta isomerism at the hexahistidine coordination site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, HEME C, ... | | Authors: | Tezcan, F.A, Kakkis, A. | | Deposit date: | 2020-05-29 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9987 Å) | | Cite: | Metal-Templated Design of Chemically Switchable Protein Assemblies with High-Affinity Coordination Sites.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7UI4
 
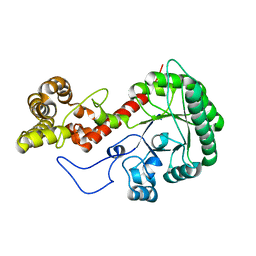 | |
8OJX
 
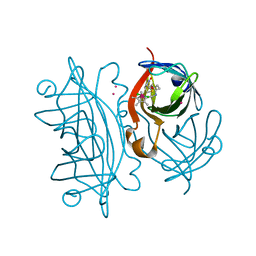 | | Streptavidin S112YK121E artificial metalloenzyme for carboamination | | Descriptor: | IRIDIUM ION, Streptavidin, trichloro{(1,2,3,4,5-eta)-1,2,3,4-tetramethyl-5-[2-({5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)ethyl]cyclopentadienyl}rhodium(1+) | | Authors: | Igareta, N.V, Ward, T.R. | | Deposit date: | 2023-03-25 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Streptavidin WT artificial metalloenzyme for carboamination
To Be Published
|
|
8EM3
 
 | |
6X83
 
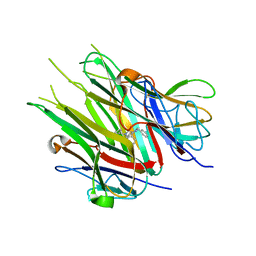 | |
5LFR
 
 | |
7UVH
 
 | | Pfs230 domain 1 bound by RUPA-32 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMMONIUM ION, ... | | Authors: | Ivanochko, D, Newton, J, Julien, J.P. | | Deposit date: | 2022-05-02 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Potent transmission-blocking monoclonal antibodies from naturally exposed individuals target a conserved epitope on Plasmodium falciparum Pfs230.
Immunity, 56, 2023
|
|
8F22
 
 | |
8EGZ
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, POTASSIUM ION, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
7UK7
 
 | |
6X9B
 
 | |
8EO7
 
 | | Crystal structure of metagenomic beta-lactamase LRA-5 Y69Q/V166E mutant at 2.15 Angstrom resolution | | Descriptor: | beta-lactamase | | Authors: | Power, P, D'Amico Gonzalez, G, Centron, D, Gutkind, G, Handelsman, J, Klinke, S. | | Deposit date: | 2022-10-02 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Playing beta-Lactamase Evolution: Metagenomic Class A beta-Lactamase LRA-5 is an Inactive Enzyme Capable of Rendering an Active beta-Lactamase by Introduction of Y69Q and V166E Substitutions
to be published
|
|
5LGV
 
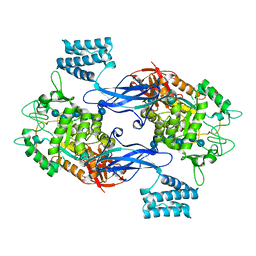 | | GlgE isoform 1 from Streptomyces coelicolor E423A mutant soaked in maltooctaose | | Descriptor: | Alpha-1,4-glucan:maltose-1-phosphate maltosyltransferase 1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Syson, K, Stevenson, C.E.M, Mia, F, Barclay, J.E, Tang, M, Gorelik, A, Rashid, A.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2016-07-08 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-bound structures and site-directed mutagenesis identify the acceptor and secondary binding sites of Streptomyces coelicolor maltosyltransferase GlgE.
J.Biol.Chem., 291, 2016
|
|
8EH0
 
 | | Engineered tyrosine synthase (TmTyrS1) derived from T. maritima TrpB with Ser bound as the amino-acrylate intermediate and complexed with quinoline N-oxide | | Descriptor: | 1,2-ETHANEDIOL, 1-oxo-1lambda~5~-quinoline, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Porter, N.J, Almhjell, P.J, Arnold, F.H. | | Deposit date: | 2022-09-13 | | Release date: | 2023-10-04 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The beta-subunit of tryptophan synthase is a latent tyrosine synthase.
Nat.Chem.Biol., 20, 2024
|
|
6XB0
 
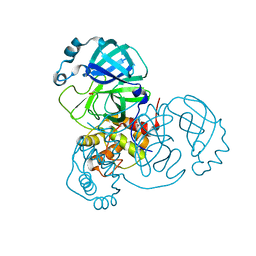 | |
8EML
 
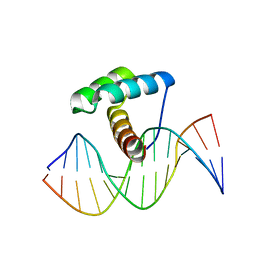 | | Crystal Structure of Gsx2 Homeodomain in Complex with DNA | | Descriptor: | DNA (5'-D(P*GP*AP*GP*CP*TP*AP*AP*TP*TP*AP*AP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*TP*TP*AP*AP*TP*TP*AP*GP*CP*TP*C)-3'), GS homeobox 2, ... | | Authors: | Webb, J.A, Kovall, R.A. | | Deposit date: | 2022-09-28 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of Gsx2 Homeodomain in Complex with DNA
To Be Published
|
|
5LKQ
 
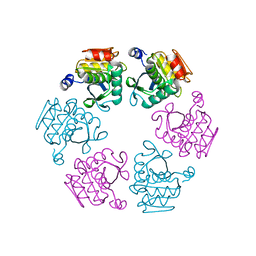 | | Protease domain of RadA | | Descriptor: | DNA repair protein RadA | | Authors: | Rapisarda, C, Fronzes, R. | | Deposit date: | 2016-07-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Bacterial RadA is a DnaB-type helicase interacting with RecA to promote bidirectional D-loop extension.
Nat Commun, 8, 2017
|
|
6XB6
 
 | |
8OIO
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 12, ... | | Authors: | Dalietou, E.V, Chen, Z, Ramdass, A.E, Manning, C, Richardson, W, Aitmakhanova, K, Platt, M, Pike, A.C.W, Fedorov, O, Brennan, P, Bullock, A.N. | | Deposit date: | 2023-03-23 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Crystal structure of the kelch domain of human KLHL12 in complex with PLEKHA4 peptide
To Be Published
|
|
8EKG
 
 | | MHETase variant Thr159Val, Met192Tyr, Tyr252Phe, Tyr503Trp | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Saunders, J.W, Frkic, R.L, Jackson, C.J. | | Deposit date: | 2022-09-20 | | Release date: | 2023-10-18 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Increasing the Soluble Expression and Whole-Cell Activity of the Plastic-Degrading Enzyme MHETase through Consensus Design.
Biochemistry, 63, 2024
|
|
8OK1
 
 | |
