6DKD
 
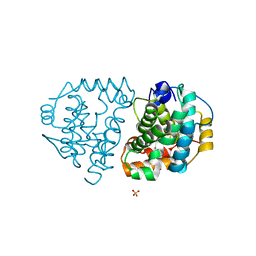 | | Yeast Ddi2 Cyanamide Hydratase | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
6DKC
 
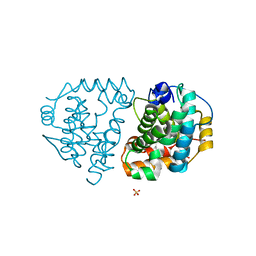 | | Yeast Ddi2 Cyanamide Hydratase, T157V mutant, apo structure | | Descriptor: | DNA damage-inducible protein, SULFATE ION, ZINC ION | | Authors: | Moore, S.A, Xiao, W, Li, J. | | Deposit date: | 2018-05-29 | | Release date: | 2019-05-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of Ddi2, a highly inducible detoxifying metalloenzyme fromSaccharomyces cerevisiae.
J.Biol.Chem., 294, 2019
|
|
1V86
 
 | | Solution structure of the ubiquitin domain from mouse D7Wsu128e protein | | Descriptor: | DNA segment, Chr 7, Wayne State University 128, ... | | Authors: | Tomizawa, T, Kigawa, T, Koshiba, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-29 | | Release date: | 2004-06-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the ubiquitin domain from mouse D7Wsu128e protein
To be Published
|
|
8TJU
 
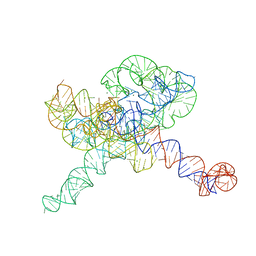 | | Tetrahymena Ribozyme scaffolded TABV xrRNA | | Descriptor: | DNA/RNA (416-MER), MAGNESIUM ION | | Authors: | Langeberg, C.J, Kieft, J.S. | | Deposit date: | 2023-07-24 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | A generalizable scaffold-based approach for structure determination of RNAs by cryo-EM.
Nucleic Acids Res., 51, 2023
|
|
1O56
 
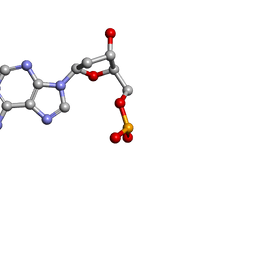 | | MOLECULAR STRUCTURE OF TWO CRYSTAL FORMS OF CYCLIC TRIADENYLIC ACID AT 1 ANGSTROM RESOLUTION | | Descriptor: | DNA (5'-CD(*AP*AP*AP*)-3') | | Authors: | Gao, Y.G, Robinson, H, Guan, Y, Liaw, Y.C, van Boom, J.H, van der Marel, G.A, Wang, A.H. | | Deposit date: | 2003-08-20 | | Release date: | 2003-08-26 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | Molecular structure of two crystal forms of cyclic triadenylic acid at 1A resolution.
J.Biomol.Struct.Dyn., 16, 1998
|
|
5Y87
 
 | |
7ECK
 
 | | RNA duplex containing C-Ag-A and U-Ag-A base pairs | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | RNA duplex containing C-Ag-A and U-Ag-A base pairs
To Be Published
|
|
7ECJ
 
 | | RNA duplex containing C-A base pairs | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3') | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | RNA duplex containing C-A base pairs
To Be Published
|
|
7ECL
 
 | | RNA duplex containing C-Ag-A and U-Ag-A base pair | | Descriptor: | DNA/RNA (5'-R(*GP*GP*AP*CP*U)-D(P*(CBR))-R(P*GP*AP*AP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Uchida, Y. | | Deposit date: | 2021-03-12 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | RNA duplex containing C-Ag-A and U-Ag-A base pair
To Be Published
|
|
5Y85
 
 | |
3RBN
 
 | | Crystal structure of MutL protein homolog 1 isoform 1 [Homo sapiens] | | Descriptor: | DNA mismatch repair protein Mlh1 | | Authors: | Dong, A, Dombrovsky, L, Wernimont, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-03-29 | | Release date: | 2011-04-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of MutL protein homolog 1 isoform 1 [Homo sapiens]
To be Published
|
|
7UU4
 
 | | Crystal structure of APOBEC3G complex with ssRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*UP*UP*U)-3'), SULFATE ION, ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU3
 
 | | Crystal structure of APOBEC3G complex with 3'overhangs RNA-Complex | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(*CP*CP*CP*AP*CP*GP*GP*GP*AP*AP*U)-3'), RNA (5'-R(*CP*CP*CP*GP*UP*GP*GP*GP*AP*AP*U)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
7UU5
 
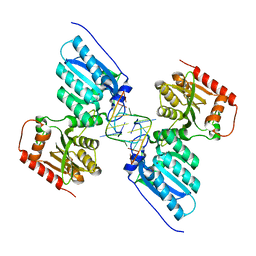 | | Crystal structure of APOBEC3G complex with 5'-Overhang dsRNA | | Descriptor: | DNA dC->dU-editing enzyme APOBEC-3G, RNA (5'-R(P*UP*AP*AP*CP*CP*GP*CP*AP*GP*CP*G)-3'), RNA (5'-R(P*UP*AP*AP*CP*GP*CP*UP*GP*CP*GP*G)-3'), ... | | Authors: | Yang, H, Li, S, Chen, X.S. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sequence-specific RNA recognition by the antiviral factor APOBEC3G.
Nat Commun, 13, 2022
|
|
241D
 
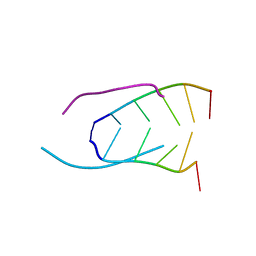 | | EXTENSION OF THE FOUR-STRANDED INTERCALATED CYTOSINE MOTIF BY ADENINE.ADENINE BASE PAIRING IN THE CRYSTAL STRUCTURE OF D(CCCAAT) | | Descriptor: | DNA (5'-D(*CP*CP*CP*AP*AP*T)-3') | | Authors: | Berger, I, Kang, C, Fredian, A, Ratliff, R, Moyzis, R, Rich, A. | | Deposit date: | 1995-12-07 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Extension of the four-stranded intercalated cytosine motif by adenine.adenine base pairing in the crystal structure of d(CCCAAT).
Nat.Struct.Biol., 2, 1995
|
|
290D
 
 | |
265D
 
 | |
271D
 
 | |
3OPW
 
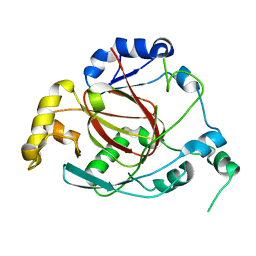 | | Crystal Structure of the Rph1 catalytic core | | Descriptor: | DNA damage-responsive transcriptional repressor RPH1 | | Authors: | Chang, Y, Wu, J, Tong, X, Zhou, J, Ding, J. | | Deposit date: | 2010-09-02 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the catalytic core of Saccharomyces cerevesiae histone demethylase Rph1: insights into the substrate specificity and catalytic mechanism
Biochem.J., 433, 2011
|
|
266D
 
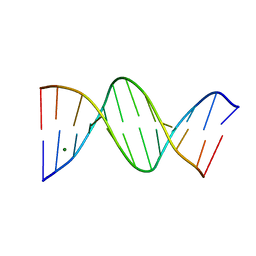 | |
291D
 
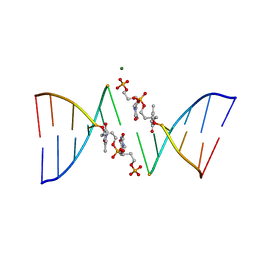 | | CRYSTAL STRUCTURES OF OLIGODEOXYRIBONUCLEOTIDES CONTAINING 6'-ALPHA-METHYL AND 6'-ALPHA-HYDROXY CARBOCYCLIC THYMIDINES | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*(T48)P*(T48)P*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Portmann, S, Altmann, K.-H, Reynes, N, Egli, M. | | Deposit date: | 1996-10-10 | | Release date: | 1996-11-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structures of Oligodeoxyribonucleotides Containing 6'-alpha-Methyl and 6'-alpha-Hydroxy Carbocyclic Thymidines
J.Am.Chem.Soc., 119, 1997
|
|
270D
 
 | |
9FCI
 
 | | USP1 bound to KSQ-4279 and ubiquitin conjugated to FANCD2 (focused refinement) | | Descriptor: | 6-(4-cyclopropyl-6-methoxy-pyrimidin-5-yl)-1-[[4-[1-propan-2-yl-4-(trifluoromethyl)imidazol-2-yl]phenyl]methyl]pyrazolo[3,4-d]pyrimidine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 2024
|
|
9FCJ
 
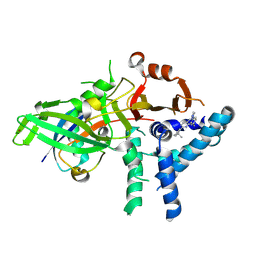 | | USP1 bound to ML323 and ubiquitin conjugated to FANCD2 (ordered subset, focused refinement) | | Descriptor: | 5-methyl-2-(2-propan-2-ylphenyl)-~{N}-[[4-(1,2,3-triazol-1-yl)phenyl]methyl]pyrimidin-4-amine, Polyubiquitin-C, Ubiquitin carboxyl-terminal hydrolase 1, ... | | Authors: | Rennie, M.L, Gundogdu, M, Walden, H. | | Deposit date: | 2024-05-15 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural and Biochemical Insights into the Mechanism of Action of the Clinical USP1 Inhibitor, KSQ-4279.
J.Med.Chem., 2024
|
|
6JMA
 
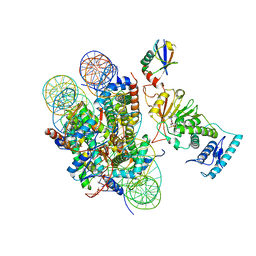 | | cryo-EM structure of DOT1L bound to H2B ubiquitinated nucleosome | | Descriptor: | DNA I&J, Histone H2A, Histone H2B 1.1, ... | | Authors: | Jang, S, Song, J.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-05-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.
Genes Dev., 33, 2019
|
|
