1ME7
 
 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP and MOA bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, MYCOPHENOLIC ACID, POTASSIUM ION, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a Catalysis-dependent
Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
1NFB
 
 | |
1MEH
 
 | |
1MEI
 
 | |
1ME8
 
 | | Inosine Monophosphate Dehydrogenase (IMPDH) From Tritrichomonas Foetus with RVP bound | | Descriptor: | INOSINE-5'-MONOPHOSPHATE DEHYDROGENASE, POTASSIUM ION, RIBAVIRIN MONOPHOSPHATE, ... | | Authors: | Prosise, G.L, Wu, J, Luecke, H. | | Deposit date: | 2002-08-08 | | Release date: | 2003-01-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Tritrichomonas foetus Inosine Monophosphate Dehydrogenase
in Complex with the Inhibitor Ribavirin Monophosphate Reveals a
Catalysis-dependent Ion-binding Site
J.Biol.Chem., 277, 2002
|
|
4Z87
 
 | | Structure of the IMP dehydrogenase from Ashbya gossypii bound to GDP | | Descriptor: | ACETATE ION, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Buey, R.M, de Pereda, J.M, Revuelta, J.L. | | Deposit date: | 2015-04-08 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases.
Nat Commun, 6, 2015
|
|
6U8N
 
 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Fully extended filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8E
 
 | |
6UDQ
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament end reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDO
 
 | | Human IMPDH2 treated with ATP, IMP, and 20 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-19 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UDP
 
 | |
6UA4
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (3/4 compressed, 1/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8R
 
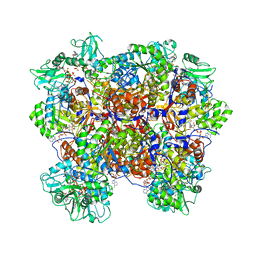 | | Human IMPDH2 treated with ATP, IMP, and NAD+. Bent (1/4 compressed, 3/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U9O
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Fully compressed filament segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-09 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UA5
 
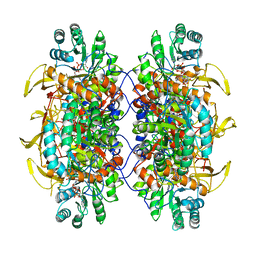 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free interfacial octamer reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6U8S
 
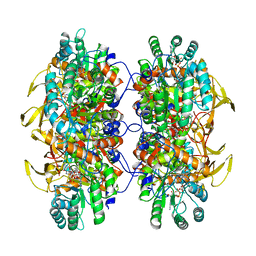 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Filament assembly interface reconstruction. | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase 2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-05 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UAJ
 
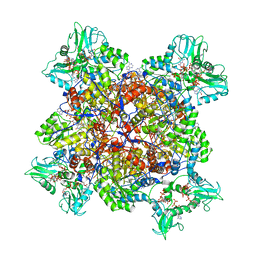 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Free canonical octamer reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
6UC2
 
 | |
6UA2
 
 | | Human IMPDH2 treated with ATP, IMP, NAD+, and 2 mM GTP. Bent (2/4 compressed, 2/4 extended) segment reconstruction. | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSINIC ACID, ... | | Authors: | Johnson, M.C, Kollman, J.M. | | Deposit date: | 2019-09-10 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structures demonstrate human IMPDH2 filament assembly tunes allosteric regulation.
Elife, 9, 2020
|
|
5TC3
 
 | | Structure of IMP dehydrogenase from Ashbya gossypii bound to ATP and GDP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fernandez-Justel, D, de Pereda, J.M, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | A nucleotide-controlled conformational switch modulates the activity of eukaryotic IMP dehydrogenases.
Sci Rep, 7, 2017
|
|
7PMZ
 
 | | Crystal structure of Streptomyces coelicolor guaB (IMP dehydrogenase) bound to ATP and ppGpp at 2.0 A resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Fernandez-Justel, D, Revuelta, J.L, Buey, R.M. | | Deposit date: | 2021-09-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Diversity of mechanisms to control bacterial GTP homeostasis by the mutually exclusive binding of adenine and guanine nucleotides to IMP dehydrogenase.
Protein Sci., 31, 2022
|
|
7PJI
 
 | |
7QBJ
 
 | | bacterial IMPDH chimera | | Descriptor: | Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QEM
 
 | | bacterial IMPDH chimera | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase | | Authors: | Labesse, G, Gelin, M, Gedeon, A, Haouz, A, Munier-Lehmann, H. | | Deposit date: | 2021-12-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
7QDX
 
 | | bacterial IMPDH chimera | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, MAGNESIUM ION | | Authors: | Labesse, G, Gelin, M, Munier-Lehmann, H, Gedeon, A, Haouz, A. | | Deposit date: | 2021-11-30 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the role of the Bateman domain at the molecular and physiological levels through engineered IMP dehydrogenases.
Protein Sci., 32, 2023
|
|
