6FTX
 
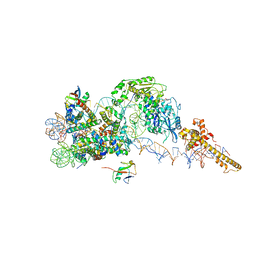 | | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromatin-remodeling ATPase, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-02-25 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
8QXB
 
 | | TDP-43 amyloid fibrils: Morphology-2 | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QXA
 
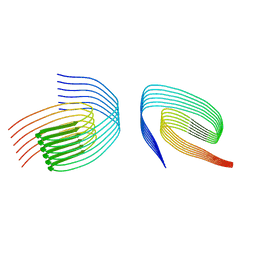 | | TDP-43 amyloid fibrils: Morphology-1b | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
8QX9
 
 | | TDP-43 amyloid fibrils: Morphology-1a | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Sharma, K, Shenoy, J, Loquet, A, Schmidt, M, Faendrich, M. | | Deposit date: | 2023-10-24 | | Release date: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Cryo-EM observation of the amyloid key structure of polymorphic TDP-43 amyloid fibrils.
Nat Commun, 15, 2024
|
|
1Y84
 
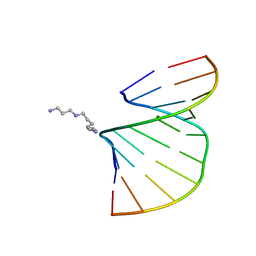 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-(imidazolyl)ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(EIT)P*AP*CP*GP*C)-3', MAGNESIUM ION, SPERMINE | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-10 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
2HYE
 
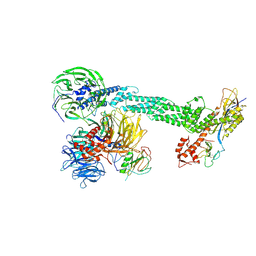 | | Crystal Structure of the DDB1-Cul4A-Rbx1-SV5V Complex | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, Nonstructural protein V, ... | | Authors: | Angers, S, Li, T, Yi, X, MacCoss, M.J, Moon, R.T, Zheng, N. | | Deposit date: | 2006-08-05 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular architecture and assembly of the DDB1-CUL4A ubiquitin ligase machinery.
Nature, 443, 2006
|
|
4A0B
 
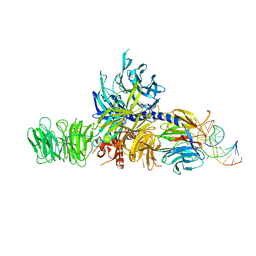 | | Structure of hsDDB1-drDDB2 bound to a 16 bp CPD-duplex (pyrimidine at D-1 position) at 3.8 A resolution (CPD 4) | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*TP*CP*CP*TP*TP*TP*CP*AP*CP*CP*C)-3', 5'-D(*DGP*GP*TP*GP*AP*AP*AP*(TTD)P*AP*GP*CP*AP*GP*DGP)-3', DNA DAMAGE-BINDING PROTEIN 1, ... | | Authors: | Scrima, A, Fischer, E.S, Iwai, S, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
1Y7F
 
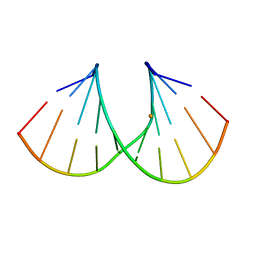 | | Crystal structure of the A-DNA GCGTAT*CGC with a 2'-O-[2-[hydroxy(methyleneamino)oxy]ethyl] Thymidine (T*) | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*(2NT)P*AP*CP*GP*C)-3'), BARIUM ION | | Authors: | Egli, M, Minasov, G, Tereshko, V, Pallan, P.S, Teplova, M, Inamati, G.B, Lesnik, E.A, Owens, S.R, Ross, B.S, Prakash, T.P, Manoharan, M. | | Deposit date: | 2004-12-08 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Influence of Stereoelectronic Effects on the Biophysical Properties of Oligonucleotides: Comprehensive Analysis of the RNA Affinity, Nuclease Resistance, and Crystal Structure of Ten 2'-O-Ribonucleic Acid Modifications.
Biochemistry, 44, 2005
|
|
2GPX
 
 | |
4PQK
 
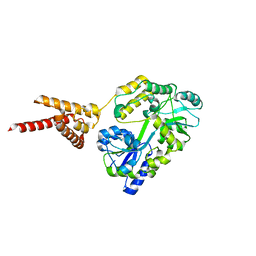 | | C-Terminal domain of DNA binding protein | | Descriptor: | Maltose ABC transporter periplasmic protein, Truncated replication protein RepA, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schumacher, M.A, Chinnam, N, Tonthat, N.K. | | Deposit date: | 2014-03-03 | | Release date: | 2014-06-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.401 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7KUI
 
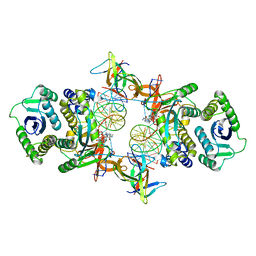 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. CIC region of a cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-25 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
7KU7
 
 | | Cryo-EM structure of Rous sarcoma virus cleaved synaptic complex (CSC) with HIV-1 integrase strand transfer inhibitor MK-2048. Cluster identified by 3-dimensional variability analysis in cryoSPARC. | | Descriptor: | (6S)-2-(3-chloro-4-fluorobenzyl)-8-ethyl-10-hydroxy-N,6-dimethyl-1,9-dioxo-1,2,6,7,8,9-hexahydropyrazino[1',2':1,5]pyrrolo[2,3-d]pyridazine-4-carboxamide, DNA (5'-D(*AP*AP*TP*GP*TP*TP*GP*TP*CP*TP*TP*AP*TP*GP*CP*AP*AP*T)-3'), DNA (5'-D(*AP*TP*TP*GP*CP*AP*TP*AP*AP*GP*AP*CP*AP*AP*CP*A)-3'), ... | | Authors: | Pandey, K.K, Bera, S, Shi, K, Aihara, H, Grandgenett, D.P. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structure of the Rous sarcoma virus octameric cleaved synaptic complex intasome.
Commun Biol, 4, 2021
|
|
9BS2
 
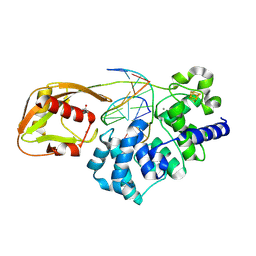 | | Glycosylase MutY variant R149Q in complex with DNA containing d(8-oxo-G) paired with a product analog (THF) to 1.51 A resolution | | Descriptor: | ACETIC ACID, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Trasvina-Arenas, C.H, Tamayo, N, Lin, W.J, Demir, M, Fisher, A.J, David, S.S, Horvath, M.P. | | Deposit date: | 2024-05-12 | | Release date: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural and functional profiling of MUTYH cancer-associated variants reveal an allosteric role for its
[4Fe-4S] cluster cofactor
To Be Published
|
|
3MU6
 
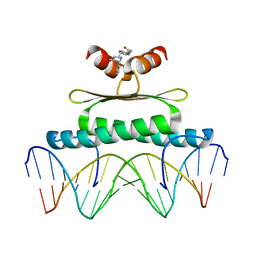 | | Inhibiting the Binding of Class IIa Histone Deacetylases to Myocyte Enhancer Factor-2 by Small Molecules | | Descriptor: | (3E)-N~8~-(2-aminophenyl)-N~1~-phenyloct-3-enediamide, DNA (5'-D(*AP*AP*AP*GP*CP*TP*AP*TP*TP*AP*TP*TP*AP*GP*CP*TP*T)-3'), DNA (5'-D(*TP*AP*AP*GP*CP*TP*AP*AP*TP*AP*AP*TP*AP*GP*CP*TP*T)-3'), ... | | Authors: | Jayathilaka, N, Han, A, Gaffney, K, Dey, R, He, J, Ye, J, Gao, T, Petasis, N.A, Chen, L. | | Deposit date: | 2010-05-01 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.434 Å) | | Cite: | Inhibition of the function of class IIa HDACs by blocking their interaction with MEF2.
Nucleic Acids Res., 40, 2012
|
|
3MX9
 
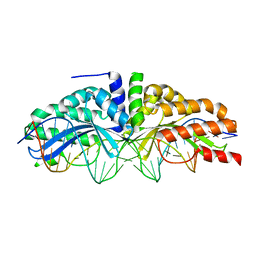 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
3MXB
 
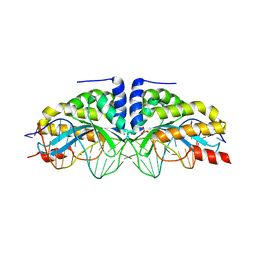 | | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus | | Descriptor: | CALCIUM ION, DNA (5'-D(*TP*CP*TP*GP*GP*CP*TP*GP*AP*GP*GP*TP*AP*CP*CP*TP*GP*AP*GP*AP*AP*CP*AP*A)-3'), DNA (5'-D(*TP*TP*GP*TP*TP*CP*TP*CP*AP*GP*GP*TP*AP*CP*CP*TP*CP*AP*GP*CP*CP*AP*GP*A)-3'), ... | | Authors: | Munoz, I.G, Prieto, J, Subramanian, S, Coloma, J, Montoya, G. | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of engineered meganuclease targeting of the endogenous human RAG1 locus.
Nucleic Acids Res., 39, 2011
|
|
5D46
 
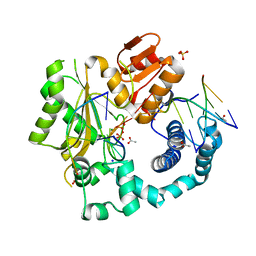 | | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(*TP*TP*TP*TP*TP*GP*C)-3'), ... | | Authors: | Loc'h, J, Rosario, S, Delarue, M. | | Deposit date: | 2015-08-07 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for a New Templated Activity by Terminal Deoxynucleotidyl Transferase: Implications for V(D)J Recombination.
Structure, 24, 2016
|
|
2GIH
 
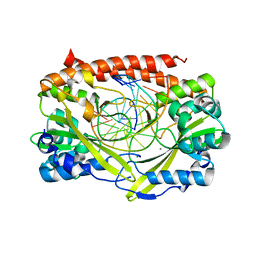 | | Q138F HincII bound to cognate DNA GTCGAC and Ca2+ | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*CP*GP*AP*CP*CP*GP*GP*C)-3', CALCIUM ION, Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K, Etzkorn, C, Chatwell, L, Bitinaite, J. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
1RDR
 
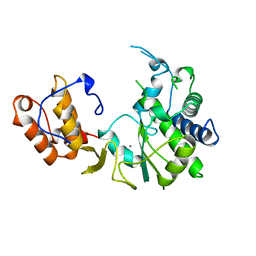 | | POLIOVIRUS 3D POLYMERASE | | Descriptor: | CALCIUM ION, POLIOVIRUS 3D POLYMERASE | | Authors: | Hansen, J, Long, A, Schultz, S. | | Deposit date: | 1998-04-28 | | Release date: | 1998-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase of poliovirus.
Structure, 5, 1997
|
|
2GII
 
 | | Q138F HincII bound to cognate DNA GTTAAC | | Descriptor: | 5'-D(*GP*CP*CP*GP*GP*TP*TP*AP*AP*CP*CP*GP*GP*C)-3', Type II restriction enzyme HincII | | Authors: | Horton, N.C, Joshi, H.K. | | Deposit date: | 2006-03-28 | | Release date: | 2006-07-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alteration of Sequence Specificity of the Type II Restriction Endonuclease HincII through an Indirect Readout Mechanism.
J.Biol.Chem., 281, 2006
|
|
4PNW
 
 | | E. coli sliding clamp in complex with (R)-6-bromo-9-(2-((S)-1-carboxy-2-phenylethylamino)-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid | | Descriptor: | (2R)-6-bromo-9-(2-{[(1S)-1-carboxy-2-phenylethyl]amino}-2-oxoethyl)-2,3,4,9-tetrahydro-1H-carbazole-2-carboxylic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Yin, Z, Oakley, A.J. | | Deposit date: | 2014-02-21 | | Release date: | 2014-03-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bacterial Sliding Clamp Inhibitors that Mimic the Sequential Binding Mechanism of Endogenous Linear Motifs.
J.Med.Chem., 58, 2015
|
|
8FNG
 
 | | Structure of E138K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNM
 
 | | Structure of G140A/Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNH
 
 | | Structure of Q148K HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-27 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
8FNP
 
 | | Structure of E138K/G140S/Q148H HIV-1 intasome with Dolutegravir bound | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, DNA (25-MER), DNA (27-MER), ... | | Authors: | Shan, Z.L, Passos, D.O, Strutzenberg, T.S, Li, M, Lyumkis, D. | | Deposit date: | 2022-12-28 | | Release date: | 2023-08-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Mechanisms of HIV-1 integrase resistance to dolutegravir and potent inhibition of drug-resistant variants.
Sci Adv, 9, 2023
|
|
