3DWC
 
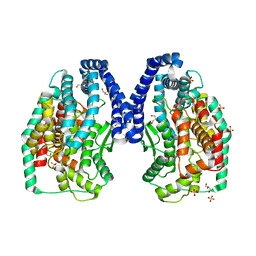 | | Trypanosoma Cruzi Metallocarboxypeptidase 1 | | Descriptor: | ALANINE, COBALT (II) ION, GLYCEROL, ... | | Authors: | Niemirowicz, G, Fernandez, D, Sola, M, Cazzulo, J.J, Aviles, F.X, Gomis-Ruth, F.X. | | Deposit date: | 2008-07-22 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular analysis of Trypanosoma cruzi metallocarboxypeptidase 1 provides insight into fold and substrate specificity
Mol.Microbiol., 70, 2008
|
|
7G3O
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-(2,3-dihydro-1H-inden-2-ylamino)-5,7-dihydropyrrolo[3,4-d]pyrimidin-6-yl]-2-[2-(1H-triazol-5-yl)ethoxy]ethanone, i.e. SMILES c12c(cccc1)CC(C2)Nc1ncc2c(n1)CN(C2)C(=O)COCCC1=CN=NN1 with IC50=0.0017376 microM | | Descriptor: | 1-{2-[(2,3-dihydro-1H-inden-2-yl)amino]-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidin-6-yl}-2-[2-(1H-1,2,3-triazol-5-yl)ethoxy]ethan-1-one, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Pinard, E, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3MEE
 
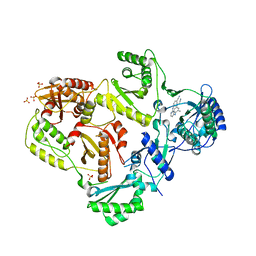 | | HIV-1 Reverse Transcriptase in Complex with TMC278 | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
1BL3
 
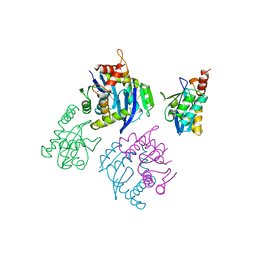 | | CATALYTIC DOMAIN OF HIV-1 INTEGRASE | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Maignan, S, Guilloteau, J.P, Zhou-Liu, Q, Clement-Mella, C, Mikol, V. | | Deposit date: | 1998-07-23 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases.
J.Mol.Biol., 282, 1998
|
|
5THR
 
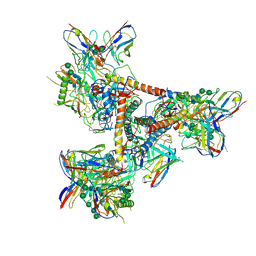 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1E2P
 
 | | Thymidine kinase, DHBT | | Descriptor: | 6-[3-HYDROXY-2-(HYDROXYMETHYL)PROPYL]-5-METHYL-2,4(1H,3H)-PYRIMIDINEDIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Schulz, G.E, Kessler, U. | | Deposit date: | 2000-05-23 | | Release date: | 2001-04-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
1E2N
 
 | | HPT + HMTT | | Descriptor: | 6-{[4-(HYDROXYMETHYL)-5-METHYL-2,6-DIOXOHEXAHYDROPYRIMIDIN-5-YL]METHYL}-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
7G2X
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-3,4,5,6-tetrahydro-1H-cyclopenta[c]pyrrole-5-carbonyl]piperidine-4-sulfonamide, i.e. SMILES C1C2=C(C[C@H]1C(=O)N1CC[C@@H](S(=O)(=O)N)CC1)CN(C2)C(=O)c1cc(nc(c1)C1CC1)OCC1CCOCC1 with IC50=0.0015731 microM | | Descriptor: | 1-(2-{2-cyclopropyl-6-[(oxan-4-yl)methoxy]pyridine-4-carbonyl}-1,2,3,4,5,6-hexahydrocyclopenta[c]pyrrole-5-carbonyl)piperidine-4-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
3CV9
 
 | | Crystal structure of vitamin D hydroxylase cytochrome P450 105A1 (R73A/R84A mutant) in complex with 1alpha,25-dihydroxyvitamin D3 | | Descriptor: | 5-{2-[1-(5-HYDROXY-1,5-DIMETHYL-HEXYL)-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE]-ETHYLIDENE}-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, Cytochrome P450-SU1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hayashi, K, Sugimoto, H, Shinkyo, R, Yamada, M, Ikeda, S, Ikushiro, S, Kamakura, M, Shiro, Y, Sakaki, T. | | Deposit date: | 2008-04-18 | | Release date: | 2008-11-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a highly active vitamin D hydroxylase from Streptomyces griseolus CYP105A1
Biochemistry, 47, 2008
|
|
3MEC
 
 | | HIV-1 Reverse Transcriptase in Complex with TMC125 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, SULFATE ION, p51 Reverse transcriptase, ... | | Authors: | Lansdon, E.B. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Etravirine (TMC125) and Rilpivirine (TMC278): Implications for Drug Design.
J.Med.Chem., 53, 2010
|
|
2JGQ
 
 | | Kinetics and structural properties of triosephosphate isomerase from Helicobacter pylori | | Descriptor: | 1-[(3-CYCLOHEXYLPROPANOYL)(2-HYDROXYETHYL)AMINO]-1-DEOXY-D-ALLITOL, PHOSPHATE ION, TRIOSEPHOSPHATE ISOMERASE | | Authors: | Chu, C.-H, Lai, Y.-J, Sun, Y.-J. | | Deposit date: | 2007-02-14 | | Release date: | 2008-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetics and Structural Properties of Triosephosphate Isomerase from Helicobacter Pylori
Proteins: Struct., Funct., Bioinf., 71, 2008
|
|
7G2P
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-[2-cyclopropyl-6-(oxan-4-ylmethoxy)pyridine-4-carbonyl]-1,3,4,6-tetrahydropyrrolo[3,4-c]pyrrole-5-carbonyl]-4-fluoropiperidine-4-sulfonamide, i.e. SMILES F[C@@]1(S(=O)(=O)N)CCN(CC1)C(=O)N1CC2=C(CN(C(=O)c3cc(C4CC4)nc(OCC4CCOCC4)c3)C2)C1 with IC50=0.00217681 microM | | Descriptor: | 1-[5-{2-cyclopropyl-6-[(oxan-4-yl)methoxy]pyridine-4-carbonyl}-3,4,5,6-tetrahydropyrrolo[3,4-c]pyrrole-2(1H)-carbonyl]-4-fluoropiperidine-4-sulfonamide, ACETATE ION, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4IKF
 
 | | PFV intasome with inhibitor MB-76 | | Descriptor: | 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3', AMMONIUM ION, ... | | Authors: | Taltynov, O, Demeulemeester, J, Desimmie, B.A, Suchaud, V, Billamboz, M, Lion, C, Bailly, F, Debyser, Z, Cotelle, P, Christ, F, Strelkov, S.V. | | Deposit date: | 2012-12-26 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | 2-Hydroxyisoquinoline-1,3(2H,4H)-diones (HIDs), novel inhibitors of HIV integrase with a high barrier to resistance.
Acs Chem.Biol., 8, 2013
|
|
1VG0
 
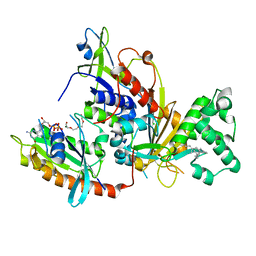 | | The crystal structures of the REP-1 protein in complex with monoprenylated Rab7 protein | | Descriptor: | CHLORIDE ION, GERAN-8-YL GERAN, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Rak, A, Pylypenko, O, Niculae, A, Pyatkov, K, Goody, R.S, Alexandrov, K. | | Deposit date: | 2004-04-22 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Rab7:REP-1 complex: insights into the mechanism of Rab prenylation and choroideremia disease
Cell(Cambridge,Mass.), 117, 2004
|
|
4AXD
 
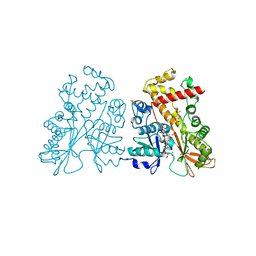 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with AMPPNP | | Descriptor: | CITRIC ACID, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
7G5H
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[(3aR,6aR)-5-(1H-benzotriazole-5-carbonyl)-1,3,3a,4,6,6a-hexahydropyrrolo[3,4-c]pyrrol-2-yl]-3-[4-(trifluoromethoxy)phenyl]propan-1-one, i.e. SMILES N1(C[C@H]2[C@H](C1)CN(C2)C(=O)c1ccc2c(c1)N=NN2)C(=O)CCc1ccc(cc1)OC(F)(F)F with IC50=0.0098589 microM | | Descriptor: | 1-[(3aR,6aS)-5-(1H-benzotriazole-5-carbonyl)hexahydropyrrolo[3,4-c]pyrrol-2(1H)-yl]-3-[4-(trifluoromethoxy)phenyl]propan-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Mattei, P, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
7G5G
 
 | | Crystal Structure of rat Autotaxin in complex with 1-[2-(cyclohexylmethyl)-1,3-dioxoisoindol-4-yl]-N-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]piperidine-4-carboxamide, i.e. SMILES N1(C(=O)c2c(C1=O)cccc2N1CCC(C(=O)NCC2=NON=C2C)CC1)CC1CCCCC1 with IC50=0.0435744 microM | | Descriptor: | 1-[2-(cyclohexylmethyl)-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl]-N-[(4-methyl-1,2,5-oxadiazol-3-yl)methyl]piperidine-4-carboxamide, CALCIUM ION, Isoform 2 of Ectonucleotide pyrophosphatase/phosphodiesterase family member 2, ... | | Authors: | Stihle, M, Benz, J, Hunziker, D, Canesso, R, Rudolph, M.G. | | Deposit date: | 2023-06-05 | | Release date: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structure of a rat Autotaxin complex
To be published
|
|
4AXE
 
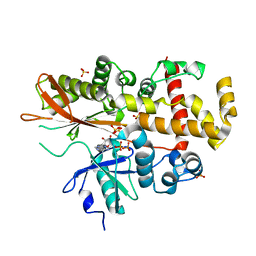 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL-PENTAKISPHOSPHATE 2-KINASE, SULFATE ION, ... | | Authors: | I Banos-Sanz, J, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2012-06-12 | | Release date: | 2012-07-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes Undergone by Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase Upon Substrate Binding: The Role of N-Lobe and Enantiomeric Substrate Preference
J.Biol.Chem., 287, 2012
|
|
1ZLF
 
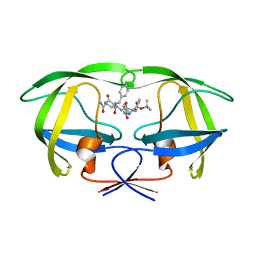 | | Crystal structure of a complex of mutant HIV-1 protease (A71V, V82T, I84V) with a hydroxyethylamine peptidomimetic inhibitor | | Descriptor: | N-{(2R,3S)-3-[(tert-butoxycarbonyl)amino]-2-hydroxy-4-phenylbutyl}-L-phenylalanyl-L-glutaminyl-L-phenylalaninamide, PROTEASE RETROPEPSIN | | Authors: | Skalova, T, Dohnalek, J, Duskova, J, Petrokova, H, Hasek, J. | | Deposit date: | 2005-05-06 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | HIV-1 protease mutations and inhibitor modifications monitored on a series of complexes. Structural basis for the effect of the A71V mutation on the active site
J.Med.Chem., 49, 2006
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
4IP7
 
 | | Structure of the S12D variant of human liver pyruvate kinase in complex with citrate and FBP. | | Descriptor: | 1,2-ETHANEDIOL, 1,6-di-O-phosphono-beta-D-fructofuranose, ADENOSINE, ... | | Authors: | Holyoak, T, Fenton, A.W. | | Deposit date: | 2013-01-09 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Energetic Coupling between an Oxidizable Cysteine and the Phosphorylatable N-Terminus of Human Liver Pyruvate Kinase.
Biochemistry, 52, 2013
|
|
5OTV
 
 | |
4B2W
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L, Gardiner, M, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-07-18 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
2Z5M
 
 | | Complex of Transportin 1 with TAP NLS, crystal form 2 | | Descriptor: | Nuclear RNA export factor 1, Transportin-1 | | Authors: | Imasaki, T, Shimizu, T, Hashimoto, H, Hidaka, Y, Kose, S, Imamoto, N, Yamada, M, Sato, M. | | Deposit date: | 2007-07-14 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for substrate recognition and dissociation by human transportin 1
Mol.Cell, 28, 2007
|
|
1QSV
 
 | | THE VEGF-BINDING DOMAIN OF FLT-1, 20 NMR STRUCTURES | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR RECEPTOR 1 | | Authors: | Starovasnik, M.A, Christinger, H.W, Wiesmann, C, Champe, M.A, de Vos, A.M, Skelton, N.J. | | Deposit date: | 1999-06-23 | | Release date: | 1999-11-10 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the VEGF-binding domain of Flt-1: comparison of its free and bound states.
J.Mol.Biol., 293, 1999
|
|
