6X5M
 
 | | Crystal structure of a stabilized PAN ENE bimolecular triplex with a GC-clamped polyA tail, in complex with Fab-BL-3,6. | | Descriptor: | Heavy chain Fab Bl-3 6, Light chain Fab BL-3 6, SULFATE ION, ... | | Authors: | Swain, M, Li, M, Wlodawer, A, Le Grice, S.F.J. | | Deposit date: | 2020-05-26 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dynamic bulge nucleotides in the KSHV PAN ENE triple helix provide a unique binding platform for small molecule ligands.
Nucleic Acids Res., 49, 2021
|
|
6WK1
 
 | | SETD3 in Complex with an Actin Peptide with His73 Replaced with Methionine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2020-04-15 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Characterization of SETD3 methyltransferase-mediated protein methionine methylation.
J.Biol.Chem., 295, 2020
|
|
4L1W
 
 | | Crystal Structuer of Human 3-alpha Hydroxysteroid Dehydrogenase Type 3 in Complex with NADP+ and Progesterone | | Descriptor: | Aldo-keto reductase family 1 member C2, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROGESTERONE, ... | | Authors: | Zhang, B, Hu, X.-J, Lin, S.-X. | | Deposit date: | 2013-06-03 | | Release date: | 2014-04-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Human 3-alpha hydroxysteroid dehydrogenase type 3 (3 alpha-HSD3): The V54L mutation restricting the steroid alternative binding and enhancing the 20 alpha-HSD activity
J.Steroid Biochem.Mol.Biol., 141, 2014
|
|
6WDP
 
 | | Interleukin 12 receptor subunit beta-1 | | Descriptor: | GLYCEROL, Interleukin-12 receptor subunit beta-1, SULFATE ION | | Authors: | Spangler, J.B, Thomas, C, Jude, K.M, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2021-12-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WDQ
 
 | | IL23/IL23R/IL12Rb1 signaling complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-12 receptor subunit beta-1, ... | | Authors: | Jude, K.M, Ely, L.K, Glassman, C.R, Thomas, C, Spangler, J.B, Lupardus, P.J, Garcia, K.C. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for IL-12 and IL-23 receptor sharing reveals a gateway for shaping actions on T versus NK cells.
Cell, 184, 2021
|
|
6WKA
 
 | | human carbonic anhydrase II bound to an inhibitor modified with azidothymidine | | Descriptor: | 3'-deoxy-3'-(4-{[(4-sulfamoylphenyl)amino]methyl}-1H-1,2,3-triazol-1-yl)thymidine, Carbonic anhydrase 2, ZINC ION | | Authors: | Peat, T.S. | | Deposit date: | 2020-04-15 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Azidothymidine "Clicked" into 1,2,3-Triazoles: First Report on Carbonic Anhydrase-Telomerase Dual-Hybrid Inhibitors.
J.Med.Chem., 63, 2020
|
|
3N4L
 
 | | BACE-1 in complex with ELN380842 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-({1-[3-(1H-pyrazol-1-yl)phenyl]cyclohexyl}amino)propyl]acetamide | | Authors: | Yao, N.H. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1RMT
 
 | | Crystal structure of AphA class B acid phosphatase/phosphotransferase complexed with adenosine. | | Descriptor: | ADENOSINE, Class B acid phosphatase, MAGNESIUM ION | | Authors: | Calderone, V, Forleo, C, Benvenuti, M, Rossolini, G.M, Thaller, M.C, Mangani, S. | | Deposit date: | 2003-11-28 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Insights in the catalytic mechanism of AphA from Escherichia coli
To be Published
|
|
6WHG
 
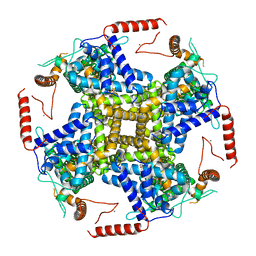 | | PI3P and calcium bound full-length TRPY1 in detergent | | Descriptor: | (2R)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1S,2S,3S,4S,5S,6R)-2,3,4,6-tetrahydroxy-5-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propan-2-yl hexadecanoate, CALCIUM ION, Calcium channel YVC1 | | Authors: | Ahmed, T, Moiseenkova-Bell, V.Y. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the ancient TRPY1 channel from Saccharomyces cerevisiae reveals mechanisms of modulation by lipids and calcium.
Structure, 30, 2022
|
|
6ZVR
 
 | | C11 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW4
 
 | | C14 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW5
 
 | | C15 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVS
 
 | | C12 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZVT
 
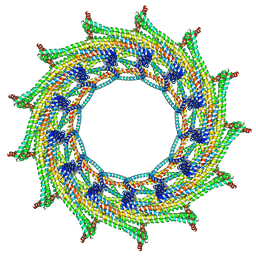 | | C13 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | Vipp1 | | Authors: | Liu, J.W, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW6
 
 | | C16 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6ZW7
 
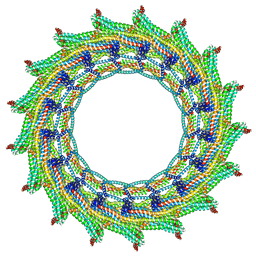 | | C17 symmetry: Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily. | | Descriptor: | vipp1 | | Authors: | Liu, J, Tassinari, M, Souza, D.P, Naskar, S, Noel, J.K, Bohuszewicz, O, Buck, M, Williams, T.A, Baum, B, Low, H.H. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (9.4 Å) | | Cite: | Bacterial Vipp1 and PspA are members of the ancient ESCRT-III membrane-remodeling superfamily.
Cell, 184, 2021
|
|
6WR1
 
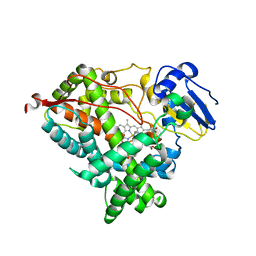 | | Human steroidogenic cytochrome P450 17A1 mutant N52Y with inhibitor abiraterone | | Descriptor: | Abiraterone, PROTOPORPHYRIN IX CONTAINING FE, Steroid 17-alpha-hydroxylase/17,20 lyase | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
6WR0
 
 | | Human steroidogenic cytochrome P450 17A1 with 3-keto-delta4-abiraterone analog | | Descriptor: | (8alpha)-17-(pyridin-3-yl)androsta-4,16-dien-3-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-04-29 | | Release date: | 2021-05-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
6X28
 
 | | Crystal structure of PT2243 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (1R)-4-(3,5-difluorophenoxy)-7-(trifluoromethyl)-2,3-dihydro-1H-inden-1-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-20 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of PT2243 bound to HIF2-PasB*:ARNT-PasB* complex
To Be Published
|
|
6X3A
 
 | |
6WW0
 
 | | Human steroidogenic cytochrome P450 17A1 with 3-keto-5alpha-abiraterone analog | | Descriptor: | (5alpha,8alpha)-17-(pyridin-3-yl)androst-16-en-3-one, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Petrunak, E.M, Bart, A.G, Scott, E.E. | | Deposit date: | 2020-05-07 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Human cytochrome P450 17A1 structures with metabolites of prostate cancer drug abiraterone reveal substrate-binding plasticity and a second binding site.
J.Biol.Chem., 299, 2023
|
|
6X37
 
 | | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 3-fluoro-5-{[(7R)-7-hydroxy-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-4-yl]oxy}benzonitrile, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of PT3245 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X39
 
 | |
6X3D
 
 | | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | (6R,7S)-4-[(3,3-difluorocyclobutyl)oxy]-6-fluoro-1-(trifluoromethyl)-6,7-dihydro-5H-cyclopenta[c]pyridin-7-ol, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-21 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PT3388 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
6X21
 
 | | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex | | Descriptor: | 1-(3-bromo-5-fluorophenoxy)-4-[(difluoromethyl)sulfonyl]-2-nitrobenzene, Aryl hydrocarbon receptor nuclear translocator, Endothelial PAS domain-containing protein 1 | | Authors: | Du, X. | | Deposit date: | 2020-05-19 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structure of PT1673 bound to HIF2a-B*:ARNT-B* complex
To Be Published
|
|
