6AS3
 
 | | Structure of a phage anti-CRISPR protein | | Descriptor: | NHis AcrE1 protein | | Authors: | Shah, M, Calmettes, C, Pawluk, A, Mejdani, M, Davidson, A.R, Maxwell, K.L, Moraes, T.F. | | Deposit date: | 2017-08-23 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Disabling a Type I-E CRISPR-Cas Nuclease with a Bacteriophage-Encoded Anti-CRISPR Protein.
MBio, 8, 2017
|
|
8CUL
 
 | | Xray ray crystal structure of OXA-24/40 in complex with CR167 | | Descriptor: | 3-({[(dihydroxyboranyl)methyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
7O2T
 
 | | PqsR (MvfR) in complex with antagonist 61 | | Descriptor: | 2-[4-[(2S)-3-(6-chloro-4-oxoquinazolin-3-yl)-2-hydroxypropoxy]phenyl]acetonitrile, LysR family transcriptional regulator | | Authors: | Emsley, J, Richardson, W. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | PqsR (MvfR) in complex with antagonist 61
To Be Published
|
|
6MA1
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor 4a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2R)-2-{[(7-chloro-2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}-2-(2-methoxyphenyl)acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
5BXO
 
 | | Human Tankyrase-2 in Complex with Macrocyclised Extended Peptide cp4n2m3 | | Descriptor: | 4,4'-propane-1,3-diylbis(1-methyl-1H-1,2,3-triazole), DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Xu, W, Fischer, G, Hyvonen, M, Itzhaki, L. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-29 | | Last modified: | 2023-03-29 | | Method: | X-RAY DIFFRACTION (1.334 Å) | | Cite: | Macrocyclized Extended Peptides: Inhibiting the Substrate-Recognition Domain of Tankyrase.
J.Am.Chem.Soc., 139, 2017
|
|
6URL
 
 | | Barrier-to-autointegration factor soaked in isopropanol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-23 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8CUM
 
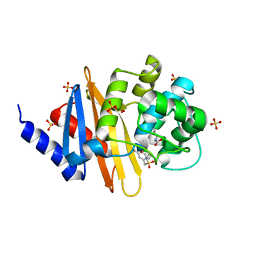 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-({[(1S)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
7WA4
 
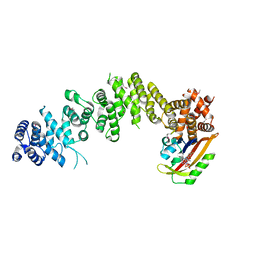 | | Crystal structure of GIGANTEA in complex with LKP2 | | Descriptor: | Adagio protein 2, FLAVIN MONONUCLEOTIDE, Protein GIGANTEA | | Authors: | Pathak, D, Dahal, P, Kwon, E, Kim, D.Y. | | Deposit date: | 2021-12-12 | | Release date: | 2022-04-27 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the regulation of blue-light receptors by GIGANTEA.
Cell Rep, 39, 2022
|
|
8CUQ
 
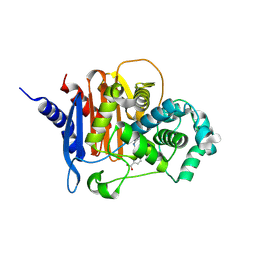 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6e | | Descriptor: | 3-[(4R)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
6URR
 
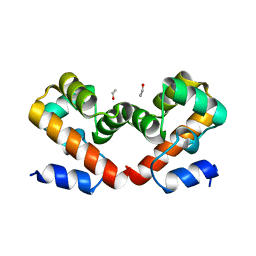 | | Barrier-to-autointegration factor soaked in Dioxane: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
7SZ9
 
 | | Crosslinked Crystal Structure of Type II Fatty Acid Synthase Ketosynthase, FabB, and C16:1-crypto Acyl Carrier Protein, AcpP | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 1, Acyl carrier protein, N~3~-{(2R)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-(2-{[(9Z)-hexadec-9-enoyl]amino}ethyl)-beta-alaninamide, ... | | Authors: | Mindrebo, J.T, Chen, A, Noel, J.P, Burkart, M.D. | | Deposit date: | 2021-11-26 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism-based cross-linking probes capture the Escherichia coli ketosynthase FabB in conformationally distinct catalytic states.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5BXU
 
 | | Human Tankyrase-2 in Complex with Macrocyclised Extended Peptide cp4n4m5 | | Descriptor: | 4,4'-pentane-1,5-diylbis(1-propyl-1H-1,2,3-triazole), CHLORIDE ION, Tankyrase-2, ... | | Authors: | Xu, W, Fischer, G, Hyvonen, M, Itzhaki, L. | | Deposit date: | 2015-06-09 | | Release date: | 2016-06-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Human Tankyrase-2 in Complex with Extended Stapled Peptide sp4n4m5
to be published
|
|
6PHJ
 
 | |
8CUO
 
 | | X-ray crystal structure of OXA-24/40 in complex with sulfonamidoboronic acid 6e | | Descriptor: | 3-({[(1R)-1-boronopropyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, SULFATE ION | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
6URV
 
 | | Crystal structure of Yellow Fever Virus NS2B-NS3 protease domain | | Descriptor: | NS2B, NS3 protease | | Authors: | Noske, G.D, Gawriljuk, V.F.O, Fernandes, R.S, Oliva, G, Godoy, A.S. | | Deposit date: | 2019-10-24 | | Release date: | 2020-01-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization and polymorphism analysis of the NS2B-NS3 protease from the 2017 Brazilian circulating strain of Yellow Fever virus.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6PHO
 
 | |
8DCM
 
 | |
6MBV
 
 | |
8CUP
 
 | | X-ray crystal structure of ADC-33 in complex with sulfonamidoboronic acid 6d | | Descriptor: | 3-[(4S)-4-ethyl-5,7,7-trihydroxy-2,2,7-trioxo-6-oxa-2lambda~6~-thia-3-aza-7lambda~5~-phospha-5-boraheptan-1-yl]benzoic acid, Beta-lactamase | | Authors: | Fernando, M.C, Wallar, B.J, Powers, R.A. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Sulfonamidoboronic Acids as "Cross-Class" Inhibitors of an Expanded-Spectrum Class C Cephalosporinase, ADC-33, and a Class D Carbapenemase, OXA-24/40: Strategic Compound Design to Combat Resistance in Acinetobacter baumannii .
Antibiotics, 12, 2023
|
|
6PHT
 
 | |
6US9
 
 | |
8CUR
 
 | | Crystal structure of Cdk2 in complex with Cyclin A inhibitor 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid | | Descriptor: | 6-[(E)-2-(4-chlorophenyl)ethenyl]-2-{[(2R)-3-(4-hydroxyphenyl)-1-methoxy-1-oxopropan-2-yl]carbamoyl}quinoline-4-carboxylic acid, Cyclin-dependent kinase 2 | | Authors: | Tripathi, S.M, Tambo, C.S, Kiss, G, Rubin, S.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biolayer Interferometry Assay for Cyclin-Dependent Kinase-Cyclin Association Reveals Diverse Effects of Cdk2 Inhibitors on Cyclin Binding Kinetics.
Acs Chem.Biol., 18, 2023
|
|
6VDB
 
 | | SETD2 in complex with a H3-variant super-substrate peptide | | Descriptor: | ALA-PRO-ARG-PHE-GLY-GLY-VAL-MET-ARG-PRO-ASN-ARG, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Beldar, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Jeltsch, A, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-24 | | Release date: | 2020-01-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence specificity analysis of the SETD2 protein lysine methyltransferase and discovery of a SETD2 super-substrate.
Commun Biol, 3, 2020
|
|
6PID
 
 | |
7O61
 
 | |
