4L7F
 
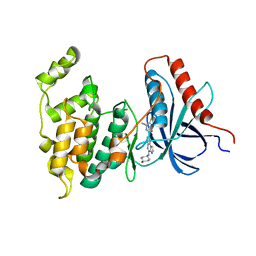 | | Co-crystal Structure of JNK1 and AX13587 | | Descriptor: | Mitogen-activated protein kinase 8, N-[1-(4-fluorophenyl)cyclopropyl]-4-[(trans-4-hydroxycyclohexyl)amino]imidazo[1,2-a]quinoxaline-8-carboxamide | | Authors: | Walter, R.L, Ranieri, G.M, Riggs, A.M, Weissig, H, Li, B, Shreder, K.R. | | Deposit date: | 2013-06-13 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Hit-to-lead optimization and kinase selectivity of imidazo[1,2-a]quinoxalin-4-amine derived JNK1 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4HAB
 
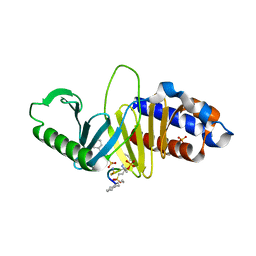 | | Crystal structure of Plk1 Polo-box domain in complex with PL-49 | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, PHOSPHATE ION, PL-49, ... | | Authors: | Lee, W.C, Song, J.H, Kim, H.Y. | | Deposit date: | 2012-09-26 | | Release date: | 2013-04-03 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of cyclic peptomer inhibitors targeting the polo-box domain of polo-like kinase 1.
Bioorg.Med.Chem., 21, 2013
|
|
5JS9
 
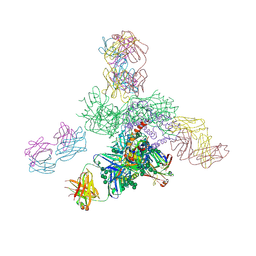 | | Uncleaved prefusion optimized gp140 trimer with an engineered 8-residue HR1 turn bound to broadly neutralizing antibodies 8ANC195 and PGT128 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2016-05-07 | | Release date: | 2016-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (6.918 Å) | | Cite: | Uncleaved prefusion-optimized gp140 trimers derived from analysis of HIV-1 envelope metastability.
Nat Commun, 7, 2016
|
|
2KOE
 
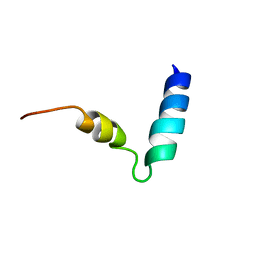 | | Human cannabinoid receptor 1 - helix 7/8 peptide | | Descriptor: | human cannabinoid receptor 1 - helix 7/8 peptide | | Authors: | Deshmukh, L, Vinogradova, O, Makriyannis, A, Tiburu, E, Tyukhtenko, S, Janero, D. | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of human cannabinoid receptor-1 helix 7/8 peptide: candidate electrostatic interactions and microdomain formation.
Biochem.Biophys.Res.Commun., 390, 2009
|
|
6BR3
 
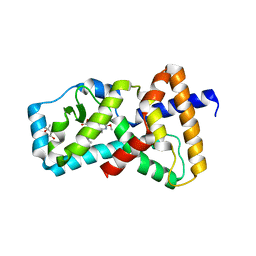 | | Structure of RORgt in complex with a novel inverse agonist TAK-828. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Nuclear receptor ROR-gamma, {cis-3-[(5R)-5-[(7-fluoro-1,1-dimethyl-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridine-6(5H)-carbonyl]cyclobutyl}acetic acid | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2017-11-29 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of [ cis-3-({(5 R)-5-[(7-Fluoro-1,1-dimethyl-2,3-dihydro-1 H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5 H)-yl}carbonyl)cyclobutyl]acetic Acid (TAK-828F) as a Potent, Selective, and Orally Available Novel Retinoic Acid Receptor-Related Orphan Receptor gamma t Inverse Agonist.
J. Med. Chem., 61, 2018
|
|
4UPU
 
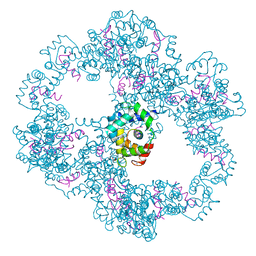 | | Crystal structure of IP3 3-K calmodulin binding region in complex with Calmodulin | | Descriptor: | CALCIUM ION, CALMODULIN, GLYCEROL, ... | | Authors: | Franco-Echevarria, E, Banos-Sanz, J.I, Monterroso, B, Round, A, Sanz-Aparicio, J, Gonzalez, B. | | Deposit date: | 2014-06-18 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | A New Calmodulin Binding Motif for Inositol 1,4,5-Trisphosphate 3-Kinase Regulation.
Biochem.J., 463, 2014
|
|
2WU4
 
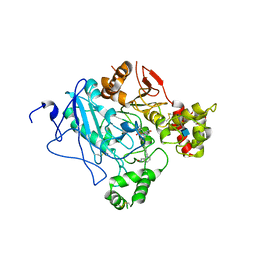 | | CRYSTAL STRUCTURE OF MOUSE ACETYLCHOLINESTERASE IN COMPLEX WITH FENAMIPHOS AND ORTHO-7 | | Descriptor: | 1,7-HEPTYLENE-BIS-N,N'-SYN-2-PYRIDINIUMALDOXIME, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Hornberg, A, Artursson, E, Warme, R, Pang, Y.-P, Ekstrom, F. | | Deposit date: | 2009-09-28 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Oxime-Bound Fenamiphos-Acetylcholinesterases: Reactivation Involving Flipping of the His447 Ring to Form a Reactive Glu334-His447-Oxime Triad.
Biochem.Pharm., 79, 2010
|
|
1DYT
 
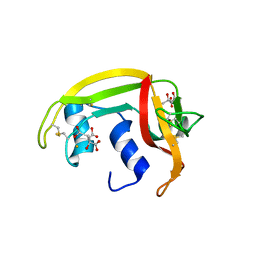 | | X-ray crystal structure of ECP (RNase 3) at 1.75 A | | Descriptor: | CITRIC ACID, EOSINOPHIL CATIONIC PROTEIN, FE (III) ION | | Authors: | Mallorqui-Fernandez, G, Pous, J, Peracaula, R, Maeda, T, Tada, H, Yamada, H, Seno, M, De Llorens, R, Gomis-Rueth, F.X, Coll, M. | | Deposit date: | 2000-02-08 | | Release date: | 2001-02-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Three-Dimensional Crystal Structure of Human Eosinophil Cationic Protein (Rnase 3) at 1.75 A Resolution.
J.Mol.Biol., 300, 2000
|
|
4JI9
 
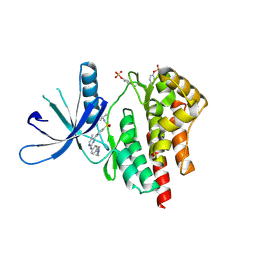 | | JAK2 kinase (JH1 domain) in complex with TG101209 | | Descriptor: | N-tert-butyl-3-[(5-methyl-2-{[4-(4-methylpiperazin-1-yl)phenyl]amino}pyrimidin-4-yl)amino]benzenesulfonamide, Tyrosine-protein kinase JAK2 | | Authors: | Eigenbrot, C, Ultsch, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 2-Amino-[1,2,4]triazolo[1,5-a]pyridines as JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
7JYD
 
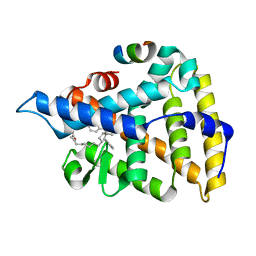 | | Human Liver Receptor Homolog-1 in Complex with 10CA and a Fragment of Tif2 | | Descriptor: | 10-[(3aR,6R,6aR)-6-hydroxy-3-phenyl-3a-(1-phenylethenyl)-1,3a,4,5,6,6a-hexahydropentalen-2-yl]decanoic acid, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2 | | Authors: | D'Agostino, E.H, Mays, S.G, Ortlund, E.A. | | Deposit date: | 2020-08-30 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tapping into a phospholipid-LRH-1 axis yields a powerful anti-inflammatory agent with in vivo activity against colitis
To Be Published
|
|
5UY8
 
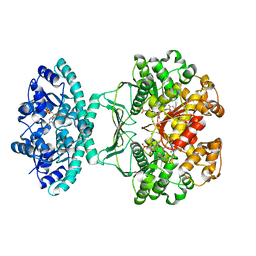 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | 5-[(5S)-5-ethyl-5-methyl-6-oxo-1,4,5,6-tetrahydropyridin-3-yl]-N-(6-fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)thiophene-2-sulfonamide, AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, ... | | Authors: | Wang, J, Wang, Y, Fales, K.R, Atwell, S, Clawson, D. | | Deposit date: | 2017-02-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
5UZ0
 
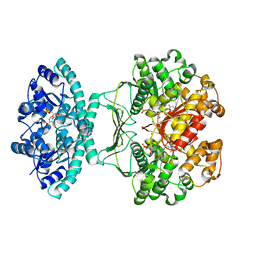 | | Crystal structure of AICARFT bound to an antifolate | | Descriptor: | AMINOIMIDAZOLE 4-CARBOXAMIDE RIBONUCLEOTIDE, Bifunctional purine biosynthesis protein PURH, MAGNESIUM ION, ... | | Authors: | Atwell, S, Wang, Y, Fales, K.R, Clawson, D, Wang, J. | | Deposit date: | 2017-02-24 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of N-(6-Fluoro-1-oxo-1,2-dihydroisoquinolin-7-yl)-5-[(3R)-3-hydroxypyrrolidin-1-yl]thiophene-2-sulfonamide (LSN 3213128), a Potent and Selective Nonclassical Antifolate Aminoimidazole-4-carboxamide Ribonucleotide Formyltransferase (AICARFT) Inhibitor Effective at Tumor Suppression in a Cancer Xenograft Model.
J. Med. Chem., 60, 2017
|
|
5AER
 
 | | Neuronal calcium sensor-1 (NCS-1)from Rattus norvegicus complex with D2 dopamine receptor peptide from Homo sapiens | | Descriptor: | CALCIUM ION, D(2) DOPAMINE RECEPTOR, NEURONAL CALCIUM SENSOR 1, ... | | Authors: | Saleem, M, Karuppiah, V, Pandalaneni, S, Burgoyne, R, Derrick, J.P, Lian, L.Y. | | Deposit date: | 2015-01-08 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Neuronal Calcium Sensor-1 Binds the D2 Dopamine Receptor and G-Protein-Coupled Receptor Kinase 1 (Grk1) Peptides Using Different Modes of Interactions.
J.Biol.Chem., 290, 2015
|
|
3HKN
 
 | | Human carbonic anhydrase II in complex with (2,3,4,6-Tetra-O-acetyl-beta-D-galactopyranosyl) -(1-4)-1,2,3,6-tetra-O-acetyl-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | 2,3,4,6-tetra-O-acetyl-beta-D-galactopyranose-(1-4)-(1S)-2,3,6-tri-O-acetyl-1,5-anhydro-1-sulfamoyl-D-glucitol, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
1JCJ
 
 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
3HKT
 
 | | Human carbonic anhydrase II in complex with alpha-D-Glucopyranosyl-(1->4)-1-thio-beta-D-glucopyranosylsulfonamide | | Descriptor: | Carbonic anhydrase 2, ZINC ION, alpha-D-galactopyranose-(1-4)-(1S)-1,5-anhydro-1-sulfamoyl-D-galactitol | | Authors: | Paul, B, Poulsen, S.-A, Hofmann, A. | | Deposit date: | 2009-05-25 | | Release date: | 2009-10-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | S-glycosyl primary sulfonamides--a new structural class for selective inhibition of cancer-associated carbonic anhydrases.
J.Med.Chem., 52, 2009
|
|
2V8F
 
 | | Mouse Profilin IIa in complex with a double repeat from the FH1 domain of mDia1 | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, PROFILIN-2, ... | | Authors: | Kursula, P, Kursula, I, Downer, J, Witke, W, Wilmanns, M. | | Deposit date: | 2007-08-07 | | Release date: | 2007-12-18 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Resolution Structural Analysis of Mammalian Profilin 2A Complex Formation with Two Physiological Ligands: The Formin Homology 1 Domain of Mdia1 and the Proline-Rich Domain of Vasp.
J.Mol.Biol., 375, 2008
|
|
3R4P
 
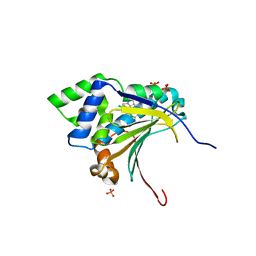 | | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide | | Descriptor: | 2-amino-4-{2,4-dichloro-6-[2-(1H-pyrazol-1-yl)ethoxy]phenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide, Heat shock protein HSP 90-alpha, PHOSPHATE ION | | Authors: | Gajiwala, K.S. | | Deposit date: | 2011-03-17 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Optimization of potent, selective, and orally bioavailable pyrrolodinopyrimidine-containing inhibitors of heat shock protein 90. Identification of development candidate 2-amino-4-{4-chloro-2-[2-(4-fluoro-1H-pyrazol-1-yl)ethoxy]-6-methylphenyl}-N-(2,2-difluoropropyl)-5,7-dihydro-6H-pyrrolo[3,4-d]pyrimidine-6-carboxamide.
J.Med.Chem., 54, 2011
|
|
3BF5
 
 | |
4HCI
 
 | | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis | | Descriptor: | Cupredoxin 1, GLYCEROL | | Authors: | Kim, Y, Maltseva, N, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-30 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Uncharacterized Cupredoxin-like Domain Protein Cupredoxin_1 from Bacillus anthracis
To be Published
|
|
2WKJ
 
 | | Crystal structure of the E192N mutant of E. Coli N-acetylneuraminic acid lyase in complex with pyruvate at 1.45A resolution in space group P212121 | | Descriptor: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, PYRUVIC ACID | | Authors: | Campeotto, I, Carr, S.B, Trinh, C.H, Nelson, A.S, Berry, A, Phillips, S.E.V, Pearson, A.R. | | Deposit date: | 2009-06-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of an Escherichia coli N-acetyl-D-neuraminic acid lyase mutant, E192N, in complex with pyruvate at 1.45 angstrom resolution.
Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun., 65, 2009
|
|
5UPI
 
 | |
3FBZ
 
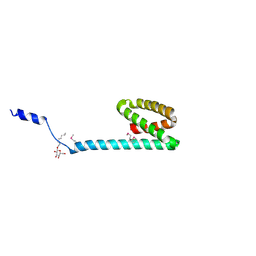 | | Crystal structure of ORF140 of the archaeal virus Acidianus Filamentous Virus 1 (AFV1) | | Descriptor: | CHLORIDE ION, Putative uncharacterized protein, octyl beta-D-glucopyranoside | | Authors: | Goulet, A, Prangishvili, D, van Tilbeurgh, H, Campanacci, V, Cambillau, C. | | Deposit date: | 2008-11-20 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6BHV
 
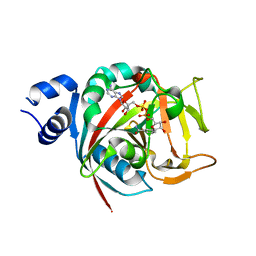 | | Human PARP-1 bound to NAD+ analog benzamide adenine dinucleotide (BAD) | | Descriptor: | Poly [ADP-ribose] polymerase 1, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl [(2R,3S,4R,5S)-5-(3-carbamoylphenyl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl dihydrogen diphosphate (non-preferred name) | | Authors: | Pascal, J.M, Langelier, M.F. | | Deposit date: | 2017-10-31 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NAD+analog reveals PARP-1 substrate-blocking mechanism and allosteric communication from catalytic center to DNA-binding domains.
Nat Commun, 9, 2018
|
|
2WLT
 
 | |
