4RD9
 
 | |
4AAZ
 
 | | X-ray structure of Nicotiana alata Defensin 1 NaD1 | | Descriptor: | 1,2-ETHANEDIOL, FLOWER-SPECIFIC DEFENSIN, PHOSPHATE ION | | Authors: | Lay, F.T, Mills, G.D, Hulett, M.D, Kvansakul, M. | | Deposit date: | 2011-12-06 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dimerization of Plant Defensin Nad1 Enhances its Antifungal Activity.
J.Biol.Chem., 287, 2012
|
|
5T5T
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-08-31 | | Release date: | 2017-03-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Selective Activation of AMPK beta 1-Containing Isoforms Improves Kidney Function in a Rat Model of Diabetic Nephropathy.
J. Pharmacol. Exp. Ther., 361, 2017
|
|
5A5P
 
 | | Crystal structure of human ATAD2 bromodomain in complex with 8-2-(dimethylamino)ethylamino-3-methyl-1,2-dihydroquinolin-2-one | | Descriptor: | 1,2-ETHANEDIOL, 8-{[2-(dimethylamino)ethyl]amino}-3-methyl-1,2-dihydroquinolin-2-one, ATPASE FAMILY AAA DOMAIN-CONTAINING PROTEIN 2, ... | | Authors: | Chung, C, Bamborough, P, Demont, E. | | Deposit date: | 2015-06-20 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-Based Discovery of Low-Micromolar Atad2 Bromodomain Inhibitors.
J.Med.Chem., 58, 2015
|
|
5DH4
 
 | | PDE10 complexed with 5-chloro-N-[(2,4-dimethylthiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine | | Descriptor: | 3-(1-hydroxy-2-methylpropan-2-yl)-5-phenyl-3,5-dihydro-1H-imidazo[4,5-c][1,8]naphthyridine-2,4-dione, 5-chloro-N-[(2,4-dimethyl-1,3-thiazol-5-yl)methyl]pyrazolo[1,5-a]pyrimidin-7-amine, MAGNESIUM ION, ... | | Authors: | Yan, Y. | | Deposit date: | 2015-08-29 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of pyrazolopyrimidine phosphodiesterase 10A inhibitors for the treatment of schizophrenia.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4QRE
 
 | |
5XC2
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
4B9Y
 
 | | Crystal Structure of Apo Agd31B, alpha-transglucosylase in Glycoside Hydrolase Family 31 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA-GLUCOSIDASE, PUTATIVE, ... | | Authors: | Larsbrink, J, Izumi, A, Hemsworth, G.R, Davies, G.J, Brumer, H. | | Deposit date: | 2012-09-09 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Enzymology of Cellvibrio Japonicus Agd31B Reveals Alpha-Transglucosylase Activity in Glycoside Hydrolase Family 31
J.Biol.Chem., 287, 2012
|
|
6B6U
 
 | | Pyruvate Kinase M2 mutant - S437Y | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Srivastava, D, Dey, M. | | Deposit date: | 2017-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Investigation of a Dimeric Variant of Pyruvate Kinase Muscle Isoform 2.
Biochemistry, 56, 2017
|
|
3CXQ
 
 | | Crystal structure of human glucosamine 6-phosphate N-acetyltransferase 1 bound to GlcN6P | | Descriptor: | 2-amino-2-deoxy-6-O-phosphono-alpha-D-glucopyranose, Glucosamine 6-phosphate N-acetyltransferase | | Authors: | Wang, J, Liu, X, Li, L.-F, Su, X.-D. | | Deposit date: | 2008-04-25 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Acceptor substrate binding revealed by crystal structure of human glucosamine-6-phosphate N-acetyltransferase 1
Febs Lett., 582, 2008
|
|
6T5A
 
 | |
4Y79
 
 | | Factor Xa complex with GTC000406 | | Descriptor: | (E)-2-(4-chlorophenyl)-N-{(3S)-1-[(2S)-1-(morpholin-4-yl)-1-oxopropan-2-yl]-2-oxopyrrolidin-3-yl}ethenesulfonamide, CALCIUM ION, Coagulation factor X, ... | | Authors: | Convery, M.A, Young, R.J, Senger, S, Hamblin, J.N, Chan, C, Toomey, J.R, Watson, N.S. | | Deposit date: | 2015-02-13 | | Release date: | 2015-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Factor Xa inhibitors: S1 binding interactions of a series of N-{(3S)-1-[(1S)-1-methyl-2-morpholin-4-yl-2-oxoethyl]-2-oxopyrrolidin-3-yl}sulfonamides.
J. Med. Chem., 50, 2007
|
|
1K1T
 
 | | Combining Mutations in HIV-1 Protease to Understand Mechanisms of Resistance | | Descriptor: | N-[(2R)-2-({N~5~-[amino(iminio)methyl]-L-ornithyl-L-valyl}amino)-4-methylpentyl]-L-phenylalanyl-L-alpha-glutamyl-L-alanyl-L-norleucinamide, PROTEASE RETROPEPSIN, SULFATE ION | | Authors: | Mahalingam, B, Boross, P, Wang, Y.-F, Louis, J.M, Fischer, C, Tozser, J, Harrison, R.W, Weber, I.T. | | Deposit date: | 2001-09-25 | | Release date: | 2002-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combining mutations in HIV-1 protease to understand mechanisms of resistance.
Proteins, 48, 2002
|
|
3D0L
 
 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 | | Descriptor: | 2F5 heavy chain, 2F5 light chain, GLYCEROL, ... | | Authors: | Bryson, S, Julien, J.P, Pai, E.F. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
6TCC
 
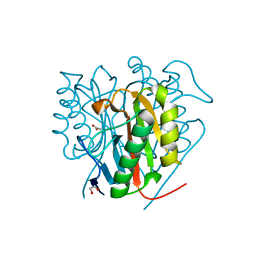 | | Crystal structure of Salmo salar RidA-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ricagno, S, Visentin, C, Di Pisa, F, Digiovanni, S, Oberti, L, Degani, G, Popolo, L, Bartorelli, A. | | Deposit date: | 2019-11-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Two novel fish paralogs provide insights into the Rid family of imine deaminases active in pre-empting enamine/imine metabolic damage.
Sci Rep, 10, 2020
|
|
3LPF
 
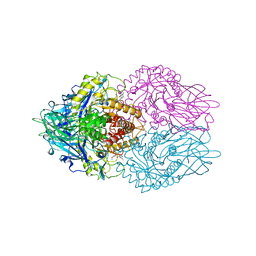 | | Structure of E. coli beta-Glucuronidase bound with a novel, potent inhibitor 1-((6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl)-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea | | Descriptor: | 1-[(6,7-dimethyl-2-oxo-1,2-dihydroquinolin-3-yl)methyl]-1-(2-hydroxyethyl)-3-(3-methoxyphenyl)thiourea, Beta-glucuronidase | | Authors: | Wallace, B.D, Redinbo, M.R. | | Deposit date: | 2010-02-05 | | Release date: | 2010-11-17 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Alleviating cancer drug toxicity by inhibiting a bacterial enzyme.
Science, 330, 2010
|
|
1NTV
 
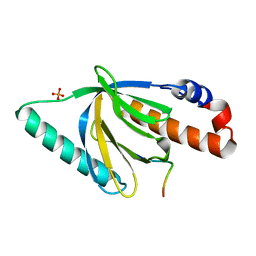 | | Crystal Structure of the Disabled-1 (Dab1) PTB domain-ApoER2 peptide complex | | Descriptor: | Apolipoprotein E Receptor-2 peptide, Disabled homolog 1, PHOSPHATE ION | | Authors: | Stolt, P.C, Jeon, H, Song, H.K, Herz, J, Eck, M.J, Blacklow, S.C. | | Deposit date: | 2003-01-30 | | Release date: | 2003-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Origins of Peptide Selectivity and Phosphoinositide Binding Revealed by Structures of Disabled-1 PTB Domain Complexes
Structure, 11, 2003
|
|
5KKW
 
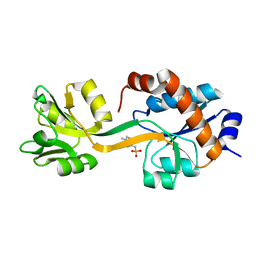 | |
5A2O
 
 | |
9CU6
 
 | | LJF-034 Fab in complex with HIV Env JRFL NFL TD CC3+ trimer and 35O22 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ozorowski, G, Ward, A.B. | | Deposit date: | 2024-07-25 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Vaccination of nonhuman primates elicits a broadly neutralizing antibody lineage targeting a quaternary epitope on the HIV-1 Env trimer.
Immunity, 58, 2025
|
|
9CU5
 
 | | LJF-085 Fab in complex with HIV Env JRFL NFL TD CC3+ trimer and 35O22 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 heavy chain Fv, ... | | Authors: | Ozorowski, G, Ward, A.B. | | Deposit date: | 2024-07-25 | | Release date: | 2025-05-21 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Vaccination of nonhuman primates elicits a broadly neutralizing antibody lineage targeting a quaternary epitope on the HIV-1 Env trimer.
Immunity, 58, 2025
|
|
3SRG
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 in complex with 2-hydroxyquinoline | | Descriptor: | BROMIDE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
4AUK
 
 | | Crystal structure of C2498 2'-O-ribose methyltransferase RlmM from Escherichia coli | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Punekar, A.S, Shepherd, T.R, Liljeruhm, J, Forster, A.C, Selmer, M. | | Deposit date: | 2012-05-18 | | Release date: | 2012-08-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Rlmm, the 2'O-Ribose Methyltransferase for C2498 of Escherichia Coli 23S Rrna.
Nucleic Acids Res., 40, 2012
|
|
3CU3
 
 | |
8UX8
 
 | | E. coli 70S ribosome with unmodified Lys-tRNAPro(GGG) bound to slippery P-site CCC-C codon in the +1 mRNA reading frame | | Descriptor: | 16S ribosomal RNA, 23S rRNA, 30S ribosomal protein S11, ... | | Authors: | Kimbrough, E.M, Dunham, C.M, Nguyen, H.A. | | Deposit date: | 2023-11-09 | | Release date: | 2025-07-09 | | Last modified: | 2025-08-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An RNA modification prevents extended codon-anticodon interactions from facilitating +1 frameshifting.
Nat Commun, 16, 2025
|
|
