7N4N
 
 | | BACE-2 in complex with ligand 36 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, N-{3-[(2S,5R)-6-amino-2-(fluoromethyl)-5-(methanesulfonyl)-5-methyl-2,3,4,5-tetrahydropyridin-2-yl]-4-fluorophenyl}-6-methoxypyrimidine-4-carboxamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7N66
 
 | | BACE-1 in complex with ligand 12 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
8QBX
 
 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
1EAJ
 
 | |
2Y7E
 
 | |
2YWP
 
 | | Crystal Structure of CHK1 with a Urea Inhibitor | | Descriptor: | 1-(5-CHLORO-2,4-DIMETHOXYPHENYL)-3-(5-CYANOPYRAZIN-2-YL)UREA, Serine/threonine-protein kinase Chk1 | | Authors: | Park, C. | | Deposit date: | 2007-04-21 | | Release date: | 2007-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and biological evaluation of 1-(2,4,5-trisubstituted phenyl)-3-(5-cyanopyrazin-2-yl)ureas as potent Chk1 kinase inhibitors
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5ELO
 
 | |
1ENP
 
 | |
1E9N
 
 | | A second divalent metal ion in the active site of a new crystal form of human apurinic/apyrimidinic endonuclease, Ape1, and its implications for the catalytic mechanism | | Descriptor: | DNA-(APURINIC OR APYRIMIDINIC SITE) LYASE, LEAD (II) ION | | Authors: | Beernink, P.T, Segelke, B.W, Rupp, B. | | Deposit date: | 2000-10-24 | | Release date: | 2001-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two Divalent Metal Ions in the Active Site of a New Crystal Form of Human Apurinic/Apyrimidinic Endonuclease, Ape1: Implications for the Catalytic Mechanism
J.Mol.Biol., 307, 2001
|
|
2Y7D
 
 | |
1F05
 
 | |
7NKE
 
 | | Crystal structure of human RXRalpha ligand binding domain in complex with 2,4-di-tert-butylphenol and a coactivator fragment | | Descriptor: | 2,4-di~{tert}-butylphenol, FORMIC ACID, Nuclear receptor coactivator 2, ... | | Authors: | Carivenc, C, Bourguet, W. | | Deposit date: | 2021-02-17 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | 2,4-Di-tert-butylphenol Induces Adipogenesis in Human Mesenchymal Stem Cells by Activating Retinoid X Receptors.
Endocrinology, 164, 2023
|
|
2M1T
 
 | |
5F3E
 
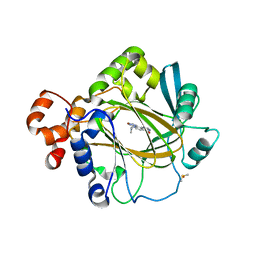 | | Crystal structure of human KDM4A in complex with compound 54a | | Descriptor: | 8-[4-[2-[4-(4-chlorophenyl)piperidin-1-yl]ethyl]pyrazol-1-yl]-3~{H}-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Le Bihan, Y.-V, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F4J
 
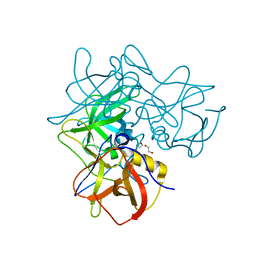 | |
2YMK
 
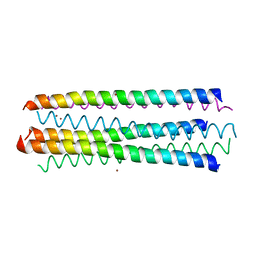 | |
7NQD
 
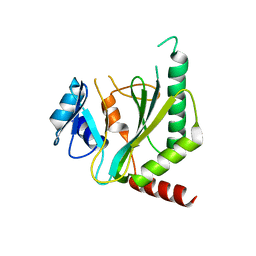 | |
7NQF
 
 | |
7NQE
 
 | |
5F09
 
 | | Structure of inactive GCPII mutant in complex with beta-citryl glutamate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Tykvart, J, Navratil, M, Pachl, P, Konvalinka, J. | | Deposit date: | 2015-11-27 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Comparison of human glutamate carboxypeptidases II and III reveals their divergent substrate specificities.
Febs J., 283, 2016
|
|
5J3X
 
 | | Structure of c-CBL Y371F | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase CBL, ZINC ION | | Authors: | Huang, D.T, Buetow, L, Dou, H. | | Deposit date: | 2016-03-31 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Casitas B-lineage lymphoma linker helix mutations found in myeloproliferative neoplasms affect conformation.
Bmc Biol., 14, 2016
|
|
5F2P
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 3. | | Descriptor: | 2-(dimethylamino)-6-methyl-pyrido[4,3-d]pyrimidin-5-one, BRD9 | | Authors: | Nar, H, Fiegen, D, Zoephel, A, Bader, G. | | Deposit date: | 2015-12-02 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F37
 
 | | Crystal structure of human KDM4A in complex with compound 58 | | Descriptor: | 1,2-ETHANEDIOL, 3H-pyrido[3,4-d]pyrimidin-4-one, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F2W
 
 | | Crystal structure of human KDM4A in complex with compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-azanyl-1,3-thiazol-4-yl)pyridine-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Le Bihan, Y.-V, Dempster, S, Westwood, I.M, van Montfort, R.L.M. | | Deposit date: | 2015-12-02 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 8-Substituted Pyrido[3,4-d]pyrimidin-4(3H)-one Derivatives As Potent, Cell Permeable, KDM4 (JMJD2) and KDM5 (JARID1) Histone Lysine Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5F4O
 
 | |
