2FWE
 
 | | crystal structure of the C-terminal domain of the electron transfer catalyst DsbD (oxidized form) | | Descriptor: | IODIDE ION, NICKEL (II) ION, SODIUM ION, ... | | Authors: | Stirnimann, C.U, Rozhkova, A, Grauschopf, U, Boeckmann, R.A, Glockshuber, R, Capitani, G, Gruetter, M.G. | | Deposit date: | 2006-02-02 | | Release date: | 2006-06-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study
J.Mol.Biol., 358, 2006
|
|
2G82
 
 | |
2GG8
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | COBALT (II) ION, METHYL N-[(2S,3R)-3-AMINO-2-HYDROXY-3-(4-METHYLPHENYL)PROPANOYL]-D-ALANYL-D-LEUCINATE, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
2GG7
 
 | | Novel bacterial methionine aminopeptidase inhibitors | | Descriptor: | 3-(5-AMINO-3-IMINO-3H-PYRAZOL-4-YLAZO)-BENZOIC ACID, COBALT (II) ION, Methionine aminopeptidase, ... | | Authors: | Evdokimov, A.G, Pokross, M.E, Walter, R.L, Mekel, M. | | Deposit date: | 2006-03-23 | | Release date: | 2006-06-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Serendipitous discovery of novel bacterial methionine aminopeptidase inhibitors.
Proteins, 66, 2007
|
|
3IMX
 
 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
3IV2
 
 | | Crystal structure of mature apo-Cathepsin L C25A mutant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cathepsin L1, GLYCEROL, ... | | Authors: | Adams-Cioaba, M.A, Krupa, J.C, Mort, J.S, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J. | | Deposit date: | 2009-08-31 | | Release date: | 2010-03-23 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the recognition and cleavage of histone H3 by cathepsin L.
Nat Commun, 2, 2011
|
|
3IC3
 
 | | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009 | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Cuff, M.E, Tesar, C, Jedrzejczak, R, Mckinlay, J.B, Harwood, C.S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-09-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a putative pyruvate dehydrogenase from the photosynthetic bacterium Rhodopseudomonas palustrus CGA009
TO BE PUBLISHED
|
|
3I8W
 
 | |
2IIT
 
 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2ID4
 
 | |
2HHA
 
 | | The structure of DPP4 in complex with an oxadiazole inhibitor | | Descriptor: | (2S,3S)-3-{3-[4-(METHYLSULFONYL)PHENYL]-1,2,4-OXADIAZOL-5-YL}-1-OXO-1-PYRROLIDIN-1-YLBUTAN-2-AMINE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G. | | Deposit date: | 2006-06-28 | | Release date: | 2006-09-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of potent, selective, and orally bioavailable oxadiazole-based dipeptidyl peptidase IV inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2IIZ
 
 | |
3J4P
 
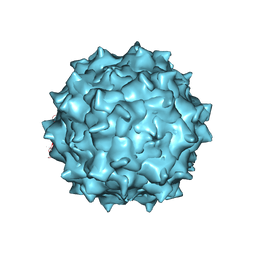 | | Electron Microscopy Analysis of a Disaccharide Analog complex Reveals Receptor Interactions of Adeno-Associated Virus | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Capsid protein VP1, MAGNESIUM ION, ... | | Authors: | Xie, Q, Chapman, M.S. | | Deposit date: | 2013-09-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Electron microscopy analysis of a disaccharide analog complex reveals receptor interactions of adeno-associated virus.
J.Struct.Biol., 184, 2013
|
|
3JY9
 
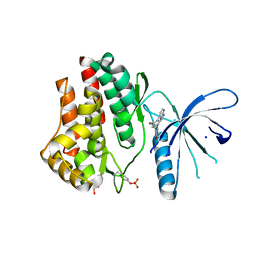 | | Janus Kinase 2 Inhibitors | | Descriptor: | (3S)-3-(4-hydroxyphenyl)-1,5-dihydro-1,5,12-triazabenzo[4,5]cycloocta[1,2,3-cd]inden-4(3H)-one, SODIUM ION, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Ledeboer, M.W, Pierce, A.C. | | Deposit date: | 2009-09-21 | | Release date: | 2009-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Janus kinase 2 inhibitors. Synthesis and characterization of a novel polycyclic azaindole.
J.Med.Chem., 52, 2009
|
|
3JPY
 
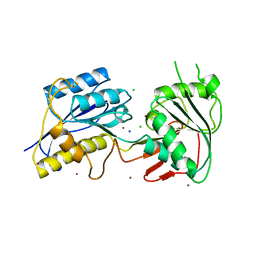 | | Crystal structure of the zinc-bound amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
3JU4
 
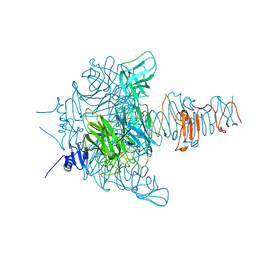 | | Crystal Structure Analysis of EndosialidaseNF at 0.98 A Resolution | | Descriptor: | CHLORIDE ION, Endo-N-acetylneuraminidase, N-acetyl-beta-neuraminic acid, ... | | Authors: | Schulz, E.C, Neuman, P, Gerardy-Schahn, R, Sheldrick, G.M, Ficner, R. | | Deposit date: | 2009-09-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Structure analysis of endosialidase NF at 0.98 A resolution.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JPW
 
 | | Crystal structure of amino terminal domain of the NMDA receptor subunit NR2B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Glutamate [NMDA] receptor subunit epsilon-2, ... | | Authors: | Karakas, E, Simorowski, N, Furukawa, H. | | Deposit date: | 2009-09-04 | | Release date: | 2009-12-08 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit.
Embo J., 28, 2009
|
|
2HYA
 
 | | HYALURONIC ACID, MOLECULAR CONFORMATIONS AND INTERACTIONS IN TWO SODIUM SALTS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranuronic acid, SODIUM ION | | Authors: | Arnott, S. | | Deposit date: | 1977-11-20 | | Release date: | 1980-03-28 | | Last modified: | 2024-02-21 | | Method: | FIBER DIFFRACTION (3 Å) | | Cite: | Hyaluronic acid: molecular conformations and interactions in two sodium salts.
J.Mol.Biol., 95, 1975
|
|
2GIN
 
 | |
2HE4
 
 | | The crystal structure of the second PDZ domain of human NHERF-2 (SLC9A3R2) interacting with a mode 1 PDZ binding motif | | Descriptor: | 1,2-ETHANEDIOL, Na(+)/H(+) exchange regulatory cofactor NHE-RF2 | | Authors: | Papagrigoriou, E, Elkins, J.M, Berridge, G, Gileady, O, Colebrook, S, Gileadi, C, Salah, E, Savitsky, P, Pantic, N, Gorrec, F, Bunkoczi, G, Weigelt, J, Arrowsmith, C, Sundstrom, M, Edwards, A, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2GMH
 
 | | Structure of Porcine Electron Transfer Flavoprotein-Ubiquinone Oxidoreductase in Complexed with Ubiquinone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, Electron transfer flavoprotein-ubiquinone oxidoreductase, ... | | Authors: | Zhang, J, Frerman, F.E, Kim, J.-J.P. | | Deposit date: | 2006-04-06 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of electron transfer flavoprotein-ubiquinone oxidoreductase and electron transfer to the mitochondrial ubiquinone pool.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3HWE
 
 | |
2GKU
 
 | | Monomeric human telomere DNA tetraplex with 3+1 strand fold topology, two edgewise loops and double-chain reversal loop, NMR, 12 structures | | Descriptor: | Human telomere DNA | | Authors: | Luu, K.N, Phan, A.T, Kuryavyi, V.V, Lacroix, L, Patel, D.J. | | Deposit date: | 2006-04-03 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in k(+) solution: an intramolecular (3 + 1) g-quadruplex scaffold.
J.Am.Chem.Soc., 128, 2006
|
|
2KJA
 
 | | Chemical shift assignments, constraints, and coordinates for CN5 scorpion toxin | | Descriptor: | Beta-toxin Cn5 | | Authors: | Prochnicka-Chalufour, A, Corzo, G, Garcia, B.I, Possani, L.D, Delepierre, M. | | Deposit date: | 2009-05-26 | | Release date: | 2009-10-06 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Cn5, a crustacean toxin found in the venom of the scorpions Centruroides noxius and Centruroides suffusus suffusus.
Biochim.Biophys.Acta, 1794, 2009
|
|
2KF7
 
 | | Structure of a two-G-tetrad basket-type intramolecular G-quadruplex formed by human telomeric repeats in K+ solution (with G7-to-BRG substitution) | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Lim, K.W, Amrane, S, Bouaziz, S, Xu, W, Mu, Y, Patel, D.J, Luu, K.N, Phan, A.T. | | Deposit date: | 2009-02-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers
J.Am.Chem.Soc., 131, 2009
|
|
