7MW4
 
 | |
5HGP
 
 | |
7MW6
 
 | |
7X6T
 
 | | Discovery of Selective BRD4 BD1 Inhibitor Based on [1,2,4] triazolo [4,3-b] pyridazine Scaffold | | Descriptor: | (2~{R})-2-[[3-methyl-6-(2-phenoxyphenyl)-[1,2,4]triazolo[4,3-b]pyridazin-8-yl]amino]propanamide, Isoform C of Bromodomain-containing protein 4 | | Authors: | Cao, D, Xiong, B. | | Deposit date: | 2022-03-08 | | Release date: | 2023-03-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Discovery of Selective BRD4 BD1 Inhibitor Based on [1,2,4] triazolo [4,3-b] pyridazine Scaffold
To Be Published
|
|
5KM5
 
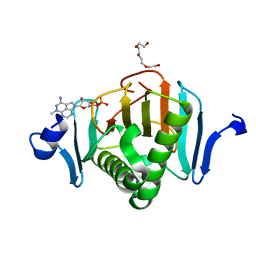 | | Human Histidine Triad Nucleotide Binding Protein 2 (hHint2) triciribine 5'-monoposphate catalytic product complex | | Descriptor: | 5-methyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-1,5-dihydro-1,4,5,6,8-pentaazaacenaphthylen-3-amine, CHLORIDE ION, Histidine triad nucleotide-binding protein 2, ... | | Authors: | Maize, K.M, Finzel, B.C. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Crystal Structure Based Guide to the Design of Human Histidine Triad Nucleotide Binding Protein 1 (hHint1) Activated ProTides.
Mol. Pharm., 14, 2017
|
|
6YAC
 
 | | Plant PSI-ferredoxin supercomplex | | Descriptor: | (1~{S})-3,5,5-trimethyl-4-[(1~{E},3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(4~{S})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-1,3,5,7,9,11,13,15,17-nonaenyl]cyclohex-3-en-1-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Caspy, I, Nelson, N, Shkolnisky, Y, Klaiman, D, Sheinker, A. | | Deposit date: | 2020-03-12 | | Release date: | 2020-09-30 | | Last modified: | 2025-10-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The structure of a triple complex of plant photosystem I with ferredoxin and plastocyanin.
Nat.Plants, 6, 2020
|
|
7BVQ
 
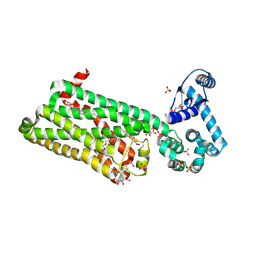 | | Structure of human beta1 adrenergic receptor bound to carazolol | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
8ORP
 
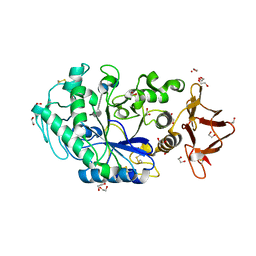 | | Crystal structure of Drosophila melanogaster alpha-amylase in complex with the inhibitor acarbose | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,2S,3S,4R,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)cyclohexyl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Aghajari, N, Haser, R. | | Deposit date: | 2023-04-16 | | Release date: | 2023-08-16 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Functional Characterization of Drosophila melanogaster alpha-Amylase.
Molecules, 28, 2023
|
|
6U63
 
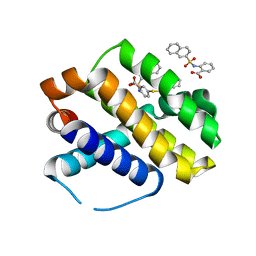 | | Mcl-1 bound to compound 17 | | Descriptor: | 2-{[(naphthalen-2-yl)sulfonyl]amino}-5-[(2-phenylethyl)sulfanyl]benzoic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Stuckey, J.A. | | Deposit date: | 2019-08-29 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery and Characterization of 2,5-Substituted Benzoic Acid Dual Inhibitors of the Anti-apoptotic Mcl-1 and Bfl-1 Proteins.
J.Med.Chem., 63, 2020
|
|
7XQE
 
 | | Crystal Structure of human RORgamma (C455E) LBD in complex with compound XY039 | | Descriptor: | 2,4-difluoro-N-(1-((4-(trifluoromethyl)benzyl)sulfonyl)-1,2,3,4-tetrahydroquinolin-7-yl)benzenesulfonamide, ETHANOL, GLYCEROL, ... | | Authors: | Wu, X, Li, C, Zhang, Y, Xu, Y. | | Deposit date: | 2022-05-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Discovery and pharmacological characterization of 1,2,3,4-tetrahydroquinoline derivatives as ROR gamma inverse agonists against prostate cancer.
Acta Pharmacol.Sin., 2024
|
|
9CFO
 
 | | Human DJ-1, 10 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
7BVT
 
 | | Crystal structure of cyclic alpha-maltosyl-1,6-maltose binding protein from Arthrobacter globiformis | | Descriptor: | Hypothetical sugar ABC-transporter sugar binding protein, alpha-D-glucopyranose-(1-6)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kohno, M, Arakawa, T, Mori, T, Nishimoto, T, Fushinobu, S. | | Deposit date: | 2020-04-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular analysis of cyclic alpha-maltosyl-(1→6)-maltose binding protein in the bacterial metabolic pathway.
Plos One, 15, 2020
|
|
3LQE
 
 | |
7X7S
 
 | | Solution structure of human adenylate kinase 1 (hAK1) | | Descriptor: | Adenylate kinase isoenzyme 1 | | Authors: | Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | ADP-Induced Conformational Transition of Human Adenylate Kinase 1 Is Triggered by Suppressing Internal Motion of alpha 3 alpha 4 and alpha 7 alpha 8 Fragments on the ps-ns Timescale.
Biomolecules, 12, 2022
|
|
7UBO
 
 | | Crystal Structure of the first bromodomain of human BRDT in complex with the inhibitor CCD-956 | | Descriptor: | Bromodomain testis-specific protein, DIMETHYL SULFOXIDE, N-[(2R)-1-(methylamino)-3-{1-[(4-methyl-2-oxo-1,2-dihydroquinolin-6-yl)acetyl]piperidin-4-yl}-1-oxopropan-2-yl]-5-phenylpyridine-2-carboxamide | | Authors: | Ta, H.M, Modukuri, R.K, Yu, Z, Tan, Z, Matzuk, M.M, Kim, C. | | Deposit date: | 2022-03-15 | | Release date: | 2022-06-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Discovery of potent BET bromodomain 1 stereoselective inhibitors using DNA-encoded chemical library selections.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8IUC
 
 | |
2V1B
 
 | |
5KX6
 
 | | The structure of Arabidopsis thaliana FUT1 Mutant R284K in complex with GDP | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Alahuhta, P.M, Lunin, V.V. | | Deposit date: | 2016-07-20 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural, mutagenic and in silico studies of xyloglucan fucosylation in Arabidopsis thaliana suggest a water-mediated mechanism.
Plant J., 91, 2017
|
|
6D40
 
 | |
2V1A
 
 | |
6CWJ
 
 | | Crystal structures of cyanuric acid hydrolase from Moorella thermoacetica complexed with 1,3-Acetone Dicarboxylic Acid | | Descriptor: | 1,3-PROPANDIOL, 3-oxopentanedioic acid, ACETATE ION, ... | | Authors: | Shi, K, Aihara, H. | | Deposit date: | 2018-03-30 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structures of Moorella thermoacetica cyanuric acid hydrolase reveal conformational flexibility and asymmetry important for catalysis.
Plos One, 14, 2019
|
|
6TKQ
 
 | | Tankyrase 2 in complex with an inhibitor (OM-2700) | | Descriptor: | Tankyrase-2, ZINC ION, ~{N}-[3-[5-(5-ethoxypyridin-2-yl)-4-phenyl-1,2,4-triazol-3-yl]cyclobutyl]-1,5-naphthyridine-4-carboxamide | | Authors: | Sowa, S.T, Lehtio, L. | | Deposit date: | 2019-11-28 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Preclinical Lead Optimization of a 1,2,4-Triazole Based Tankyrase Inhibitor.
J.Med.Chem., 63, 2020
|
|
8SPH
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
8SPI
 
 | | Crystal structure of chimeric omicron RBD (strain XBB.1.5) complexed with human ACE2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, W, Shi, K, Aihara, H, Li, F. | | Deposit date: | 2023-05-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural evolution of SARS-CoV-2 omicron in human receptor recognition.
J.Virol., 97, 2023
|
|
6E04
 
 | | sperm whale myoglobin 1-nitrosopropane | | Descriptor: | 1-nitrosopropane, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Herrera, V.E. | | Deposit date: | 2018-07-06 | | Release date: | 2019-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into Nitrosoalkane Binding to Myoglobin Provided by Crystallography of Wild-Type and Distal Pocket Mutant Derivatives.
Biochemistry, 62, 2023
|
|
