8B3F
 
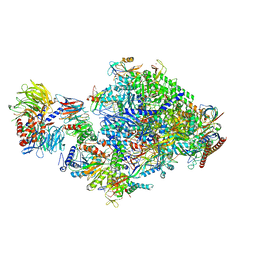 | | Pol II-CSB-CSA-DDB1-ELOF1 | | Descriptor: | DNA damage-binding protein 1, DNA excision repair protein ERCC-6, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Pol II-CSB-CSA-DDB1-ELOF1 structure.
To Be Published
|
|
9IPU
 
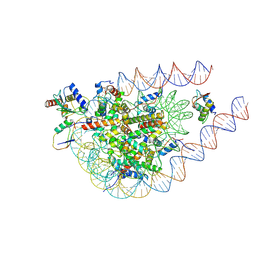 | |
8QMA
 
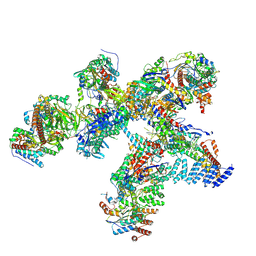 | | Structure of the plastid-encoded RNA polymerase complex (PEP) from Sinapis alba | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | do Prado, P.F.V, Ahrens, F.M, Pfannschmidt, T, Hillen, H.S. | | Deposit date: | 2023-09-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the multi-subunit chloroplast RNA polymerase.
Mol.Cell, 84, 2024
|
|
8WA1
 
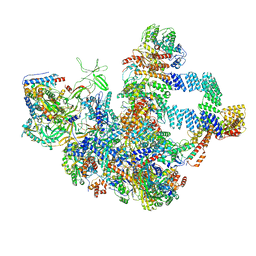 | | The cryo-EM structure of the Nicotiana tabacum PEP-PAP-TEC2 | | Descriptor: | DNA (24-mer), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structures of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8WA0
 
 | |
6EKM
 
 | |
8W9Z
 
 | | The cryo-EM structure of the Nicotiana tabacum PEP-PAP | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta'', ... | | Authors: | Wu, X.X, Zhang, Y. | | Deposit date: | 2023-09-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8CEO
 
 | |
8CEN
 
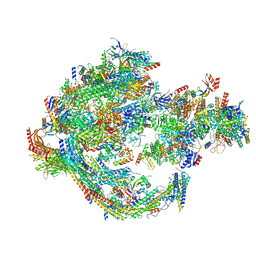 | | Yeast RNA polymerase II transcription pre-initiation complex with core Mediator | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Wang, H, Schilbach, S, Cramer, P. | | Deposit date: | 2023-02-02 | | Release date: | 2023-03-22 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Yeast PIC-Mediator structure with RNA polymerase II C-terminal domain.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4TZ4
 
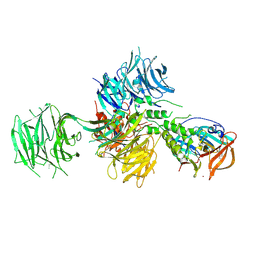 | | Crystal Structure of Human Cereblon in Complex with DDB1 and Lenalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Lenalidomide, ... | | Authors: | Chamberlain, P.P, Pagarigan, B, Delker, S, Leon, B, Riley, M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-08-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural Basis for Responsiveness to Thalidomide-Analog Drugs Defined by the Crystal Structure of the Human Cereblon:DDB1:Lenalidomide Complex
To Be Published
|
|
8OF0
 
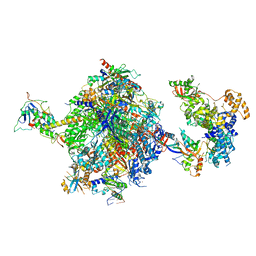 | | Structure of the mammalian Pol II-SPT6-Elongin complex, Structure 1 | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEV
 
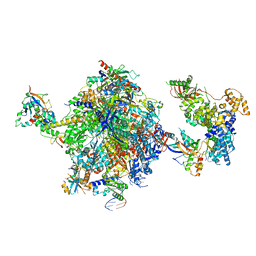 | | Structure of the mammalian Pol II-SPT6-Elongin complex, lacking ELOA latch (composite structure, structure 3) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8OEW
 
 | | Structure of the mammalian Pol II-Elongin complex, lacking the ELOA latch (composite structure, structure 2) | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Chen, Y, Kokic, G, Dienemann, C, Dybkov, O, Urlaub, H, Cramer, P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-10-18 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the transcribing RNA polymerase II-Elongin complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6DRD
 
 | | RNA Pol II(G) | | Descriptor: | DNA-directed RNA polymerase II subunit GRINL1A, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Yu, X, Jishage, M, Shi, Y, Ganesan, S, Sali, A, Chait, B.T, Asturias, F, Roeder, R.G. | | Deposit date: | 2018-06-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of Pol II(G) and molecular mechanism of transcription regulation by Gdown1.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6R75
 
 | | Crystal structure of human Ube2T E54R mutant | | Descriptor: | Ubiquitin-conjugating enzyme E2 T | | Authors: | Chaugule, V.K, Rennie, M.L, Walden, H, Arkinson, C, Kamarainen, O, Toth, R. | | Deposit date: | 2019-03-28 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric mechanism for site-specific ubiquitination of FANCD2.
Nat.Chem.Biol., 16, 2020
|
|
7YCX
 
 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
8PP7
 
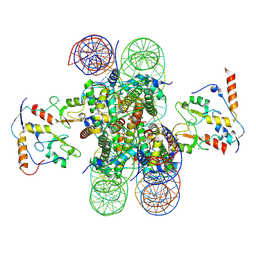 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4EXT
 
 | |
5O8K
 
 | |
8U17
 
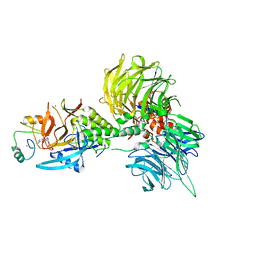 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8QU8
 
 | | PROTAC-mediated complex of KRAS with VHL/Elongin-B/Elongin-C/Cullin-2/Rbx1 | | Descriptor: | (2S,4R)-1-[(2S)-2-[4-[4-[(3S)-4-[4-[5-[(4S)-2-azanyl-3-cyano-4-methyl-6,7-dihydro-5H-1-benzothiophen-4-yl]-1,2,4-oxadiazol-3-yl]pyrimidin-2-yl]-3-methyl-1,4-diazepan-1-yl]butoxy]-1,2,3-triazol-1-yl]-3-methyl-butanoyl]-N-[(1R)-1-[4-(4-methyl-1,3-thiazol-5-yl)phenyl]-2-oxidanyl-ethyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Fischer, G, Peter, D, Arce-Solano, S. | | Deposit date: | 2023-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Targeting cancer with small-molecule pan-KRAS degraders.
Science, 385, 2024
|
|
8QH5
 
 | | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1 | | Descriptor: | DET1- and DDB1-associated protein 1, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Lee, S.-H, Sixma, T.K. | | Deposit date: | 2023-09-06 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CryoEM structure of UVSSA(VHS)-CSA-DDB1-DDA1
To Be Published
|
|
8BYA
 
 | | Cryo-EM structure of SKP1-SKP2-CKS1-CDK2-CyclinA-p27KIP1 Complex | | Descriptor: | Cyclin-A2, Cyclin-dependent kinase 2, Cyclin-dependent kinase inhibitor 1B, ... | | Authors: | Rowland, R.J, Salamina, M, Endicott, J.A, Noble, M.E. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-28 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structure of SKP1-SKP2-CKS1 in complex with CDK2-cyclin A-p27KIP1.
Sci Rep, 13, 2023
|
|
8B3I
 
 | | CRL4CSA-E2-Ub (state 2) | | Descriptor: | Cullin-4A, DNA damage-binding protein 1, DNA excision repair protein ERCC-8, ... | | Authors: | Kokic, G, Cramer, P. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | C(N)RL4CSA-E2-Ub (state 2)
To Be Published
|
|
5AVM
 
 | | Crystal structures of 5-aminoimidazole ribonucleotide (AIR) synthetase, PurM, from Thermus thermophilus | | Descriptor: | Phosphoribosylformylglycinamidine cyclo-ligase, SULFATE ION | | Authors: | Kanagawa, M, Baba, S, Watanabe, Y, Nakagawa, N, Ebihara, A, Sampei, G, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2015-06-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures and ligand binding of PurM proteins from Thermus thermophilus and Geobacillus kaustophilus
J.Biochem., 159, 2016
|
|
