2JMF
 
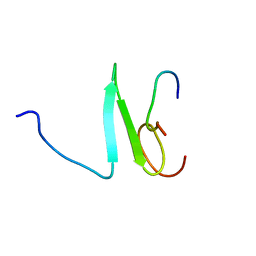 | | Solution structure of the Su(dx) WW4- Notch PY peptide complex | | Descriptor: | E3 ubiquitin-protein ligase suppressor of deltex, Neurogenic locus Notch protein | | Authors: | Avis, J.M, Blankley, R.T, Jennings, M.D, Golovanov, A.P. | | Deposit date: | 2006-11-05 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and Autoregulation of Notch Binding by Tandem WW Domains in Suppressor of Deltex
J.Biol.Chem., 282, 2007
|
|
2LOW
 
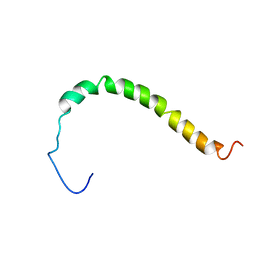 | | Solution structure of AR55 in 50% HFIP | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
2LM3
 
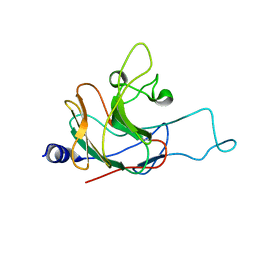 | | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain | | Descriptor: | Tripartite motif-containing protein 5 | | Authors: | Biris, N, Yang, Y, Taylor, A.B, Tomashevski, A, Guo, M, Hart, P.J, Diaz-Griffero, F, Ivanov, D.N. | | Deposit date: | 2011-11-21 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the rhesus monkey TRIM5alpha PRYSPRY domain, the HIV capsid recognition module.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2LWL
 
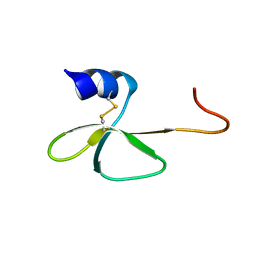 | | Structural Basis for the Interaction of Human β-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles | | Descriptor: | Beta-defensin 106 | | Authors: | de Paula, V.S, Gomes, N.S.F, Lima, L.G, Miyamoto, C.A, Monteiro, R.Q, Almeida, F.C.L, Valente, A. | | Deposit date: | 2012-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2013-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Interaction of Human beta-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles.
J.Mol.Biol., 425, 2013
|
|
2NOC
 
 | | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-10-25 | | Release date: | 2006-11-28 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Putative periplasmic protein: Northest Structural Genomics Target StR106
To be Published
|
|
2NPU
 
 | | The solution structure of the rapamycin-binding domain of mTOR (FRB) | | Descriptor: | FKBP12-rapamycin complex-associated protein | | Authors: | Veverka, V, Crabbe, T, Bird, I, Lennie, G, Muskett, F.W, Taylor, R.J, Carr, M.D. | | Deposit date: | 2006-10-30 | | Release date: | 2007-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of the interaction of mTOR with phosphatidic acid and a novel class of inhibitor: compelling evidence for a central role of the FRB domain in small molecule-mediated regulation of mTOR.
Oncogene, 27, 2008
|
|
2POA
 
 | | Schistosoma mansoni Sm14 Fatty Acid-Binding Protein: improvement of protein stability by substitution of the single Cys62 residue | | Descriptor: | 14 kDa fatty acid-binding protein | | Authors: | Ramos, C.R.R, Oyama Jr, S, Sforca, M.L, Pertinhez, T.A, Ho, P.L, Spisni, A. | | Deposit date: | 2007-04-26 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Stability improvement of the fatty acid binding protein Sm14 from S. mansoni by Cys replacement: Structural and functional characterization of a vaccine candidate.
Biochim.Biophys.Acta, 1794, 2009
|
|
2LOV
 
 | | AR55 solubilised in LPPG micelles | | Descriptor: | Apelin receptor | | Authors: | Langelaan, D.N, Rainey, J.K. | | Deposit date: | 2012-01-27 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Preserved Transmembrane Segment Topology, Structure, and Dynamics in Disparate Micellar Environments.
J Phys Chem Lett, 8, 2017
|
|
1MVK
 
 | | X-ray structure of the tetrameric mutant of the B1 domain of streptococcal protein G | | Descriptor: | Immunoglobulin G binding protein G, SULFATE ION | | Authors: | Frank, M.K, Dyda, F, Dobrodumov, A, Gronenborn, A.M. | | Deposit date: | 2002-09-25 | | Release date: | 2002-10-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Core mutations switch monomeric protein GB1 into an intertwined tetramer.
Nat.Struct.Biol., 9, 2002
|
|
1MYQ
 
 | | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction | | Descriptor: | 5'-D(*GP*GP*AP*GP*GP*AP*GP*GP*AP*GP*GP*A)-3' | | Authors: | Matsugami, A, Ouhashi, K, Kanagawa, M, Liu, H, Kanagawa, S, Uesugi, S, Katahira, M. | | Deposit date: | 2002-10-04 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction.
J.Mol.Biol., 313, 2001
|
|
1MSJ
 
 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T15V | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1NBK
 
 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-09-18 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
1NJ3
 
 | | Structure and Ubiquitin Interactions of the Conserved NZF Domain of Npl4 | | Descriptor: | NPL4, ZINC ION | | Authors: | Wang, B, Alam, S.L, Meyer, H.H, Payne, M, Stemmler, T.L, Davis, D.R, Sundquist, W.I. | | Deposit date: | 2002-12-30 | | Release date: | 2003-04-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and ubiquitin interactions of the conserved zinc finger domain of Npl4.
J.Biol.Chem., 278, 2003
|
|
1O8Z
 
 | | Solution structure of SFTI-1(6,5), an acyclic permutant of the proteinase inhibitor SFTI-1, cis-trans-trans conformer (ct-A) | | Descriptor: | CYCLIC TRYPSIN INHIBITOR | | Authors: | Marx, U.C, Craik, D.J. | | Deposit date: | 2002-12-09 | | Release date: | 2003-03-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Enzymatic Cyclization of a Potent Bowman-Birk Protease Inhibitor, Sunflower Trypsin Inhibitor-1, and Solution Structure of an Acyclic Precursor Peptide
J.Biol.Chem., 278, 2003
|
|
1OWN
 
 | | DATA3:DNA photolyase / received X-rays dose 4.8 exp15 photons/mm2 | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, PHOSPHATE ION | | Authors: | Komori, H, Adachi, S, Miki, K, Eker, A, Kort, R. | | Deposit date: | 2003-03-28 | | Release date: | 2004-04-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | DNA apophotolyase from Anacystis nidulans: 1.8 A structure, 8-HDF reconstitution and X-ray-induced FAD reduction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1MBH
 
 | | MOUSE C-MYB DNA-BINDING DOMAIN REPEAT 2 | | Descriptor: | C-MYB | | Authors: | Ogata, K, Morikawa, S, Nakamura, H, Hojo, H, Yoshimura, S, Zhang, R, Aimoto, S, Ametani, Y, Hirata, Z, Sarai, A, Ishii, S, Nishimura, Y. | | Deposit date: | 1995-05-19 | | Release date: | 1995-09-15 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb.
Nat.Struct.Biol., 2, 1995
|
|
1LVQ
 
 | | IC3 of CB1 Bound to G(alpha)i | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Ulfers, A.L, McMurry, J.L, Miller, A, Wang, L, Kendall, D.A, Mierke, D.F. | | Deposit date: | 2002-05-29 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity.
Protein Sci., 11, 2002
|
|
1NAP
 
 | |
1NEM
 
 | | Saccharide-RNA recognition in the neomycin B / RNA aptamer complex | | Descriptor: | 2,6-diamino-2,6-dideoxy-alpha-D-glucopyranose, 2,6-diamino-2,6-dideoxy-beta-L-idopyranose-(1-3)-beta-D-ribofuranose, 2-DEOXY-D-STREPTAMINE, ... | | Authors: | Jiang, L, Majumdar, A, Hu, W, Jaishree, T.J, Xu, W, Patel, D.J. | | Deposit date: | 1999-03-15 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Saccharide-RNA recognition in a complex formed between neomycin B and an RNA aptamer
Structure Fold.Des., 7, 1999
|
|
1NR3
 
 | | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212 | | Descriptor: | DNA-binding protein tfx | | Authors: | Shen, Y, Liu, G, Bhaskaran, R, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-23 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212
To be Published
|
|
1LVR
 
 | | IC3 of CB1 (L431A,A432L) Bound to G(alpha)i | | Descriptor: | Cannabinoid receptor 1 | | Authors: | Ulfers, A.L, McMurry, J.L, Miller, A, Wang, L, Kendall, D.A, Mierke, D.F. | | Deposit date: | 2002-05-29 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Cannabinoid receptor-G protein interactions: G(alphai1)-bound structures of IC3 and a mutant with altered G protein specificity.
Protein Sci., 11, 2002
|
|
1N51
 
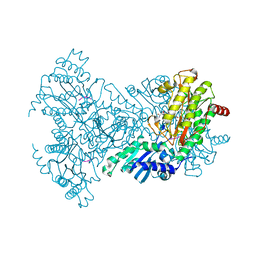 | | Aminopeptidase P in complex with the inhibitor apstatin | | Descriptor: | MANGANESE (II) ION, Xaa-Pro aminopeptidase, apstatin | | Authors: | Graham, S.C, Maher, M.J, Lee, M.H, Simmons, W.H, Freeman, H.C, Guss, J.M. | | Deposit date: | 2002-11-03 | | Release date: | 2003-12-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Escherichia coli aminopeptidase P in complex with the inhibitor apstatin.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1OH1
 
 | | Solution structure of staphostatin A form Staphylococcus aureus confirms the discovery of a novel class of cysteine proteinase inhibitors. | | Descriptor: | STAPHOSTATIN A | | Authors: | Dubin, G, Popowicz, G, Krajewski, M, Stec, J, Bochtler, M, Potempa, J, Dubin, A, Holak, T.A. | | Deposit date: | 2003-05-21 | | Release date: | 2003-11-20 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Novel Class of Cysteine Protease Inhibitors: Solution Structure of Staphostatin a from Staphylococcus Aureus
Biochemistry, 42, 2003
|
|
1MSI
 
 | |
1N17
 
 | | Structure and Dynamics of Thioguanine-modified Duplex DNA | | Descriptor: | 5'-D(*GP*CP*TP*AP*AP*GP*(S6G)P*AP*AP*AP*GP*CP*C)-3', 5'-D(*GP*GP*CP*TP*TP*TP*CP*CP*TP*TP*AP*GP*C)-3' | | Authors: | Somerville, L, Krynetski, E.Y, Krynetskaia, N.F, Beger, R.D, Zhang, W, Marhefka, C.A, Evans, W.E, Kriwacki, R.W. | | Deposit date: | 2002-10-16 | | Release date: | 2002-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of thioguanine-modified duplex DNA
J.Biol.Chem., 278, 2003
|
|
