3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
3MBX
 
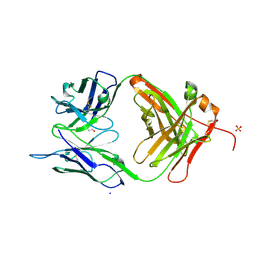 | | Crystal structure of chimeric antibody X836 | | Descriptor: | PHOSPHATE ION, SODIUM ION, X836 HEAVY CHAIN, ... | | Authors: | Teplyakov, A, Obmolova, G, Gilliland, G.L. | | Deposit date: | 2010-03-26 | | Release date: | 2010-06-30 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | On the domain pairing in chimeric antibodies.
Mol.Immunol., 47, 2010
|
|
3MF0
 
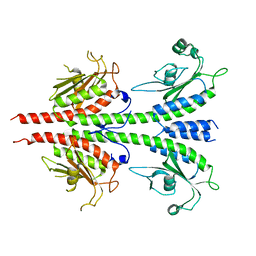 | | Crystal structure of PDE5A GAF domain (89-518) | | Descriptor: | cGMP-specific 3',5'-cyclic phosphodiesterase | | Authors: | Wang, H, Robinson, H, Ke, H. | | Deposit date: | 2010-04-01 | | Release date: | 2010-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Conformation changes, N-terminal involvement, and cGMP signal relay in the phosphodiesterase-5 GAF domain.
J.Biol.Chem., 285, 2010
|
|
2I7U
 
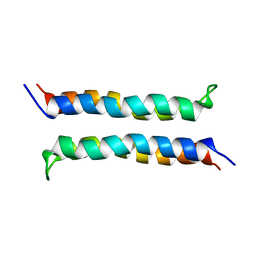 | | Structural and Dynamical Analysis of a Four-Alpha-Helix Bundle with Designed Anesthetic Binding Pockets | | Descriptor: | Four-alpha-helix bundle | | Authors: | Ma, D, Brandon, N.R, Cui, T, Bondarenko, V, Canlas, C, Johansson, J.S, Tang, P, Xu, Y. | | Deposit date: | 2006-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Four-alpha-helix bundle with designed anesthetic binding pockets. Part I: structural and dynamical analyses.
Biophys.J., 94, 2008
|
|
3MII
 
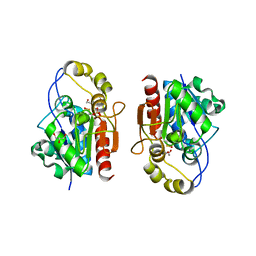 | | Crystal structure of Y0R391Cp/HSP33 from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Probable chaperone protein HSP33, SULFATE ION, ... | | Authors: | Guo, P.-C, Zhou, Y.-Y, Zhou, C.-Z, Li, W.-F. | | Deposit date: | 2010-04-10 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Hsp33/YOR391Cp from the yeast Saccharomyces cerevisiae
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2VV7
 
 | | BJFIXLH IN UNLIGANDED FERROUS FORM | | Descriptor: | CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL, ... | | Authors: | Ayers, R.A, Moffat, K. | | Deposit date: | 2008-06-04 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Changes in Quaternary Structure in the Signaling Mechanisms of Pas Domains.
Biochemistry, 47, 2008
|
|
9BRZ
 
 | | V0-only V-ATPase and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
3MW6
 
 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
9BRU
 
 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRT
 
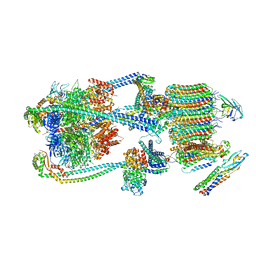 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRS
 
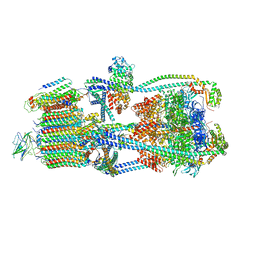 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
3MZW
 
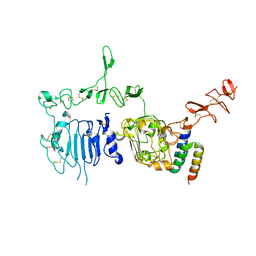 | | HER2 extracelluar region with affinity matured 3-helix affibody ZHER2:342 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G-binding protein A, ... | | Authors: | Eigenbrot, C, Ultsch, M.H. | | Deposit date: | 2010-05-13 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for high-affinity HER2 receptor binding by an engineered protein.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7JGI
 
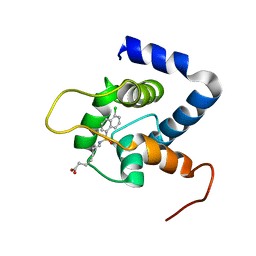 | | NMR structure of the cNTnC-cTnI chimera bound to A7 | | Descriptor: | 7-{[(5-chloronaphthalen-1-yl)sulfonyl]amino}heptanoic acid, CALCIUM ION, Troponin C, ... | | Authors: | Cai, F, Robertson, I.M, Kampourakis, T, Klein, B.A, Sykes, B.D. | | Deposit date: | 2020-07-19 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Role of Electrostatics in the Mechanism of Cardiac Thin Filament Based Sensitizers.
Acs Chem.Biol., 15, 2020
|
|
3MWN
 
 | | Structure of the Novel 14 kDa Fragment of alpha-Subunit of Phycoerythrin from the Starving Cyanobacterium Phormidium Tenue | | Descriptor: | PHYCOCYANOBILIN, PHYCOERYTHRIN | | Authors: | Soni, B.R, Hasan, M.I, Parmar, A, Ethayathulla, A.S, Kumar, R.P, Singh, N.K, Sinha, M, Kaur, P, Yadav, S, Sharma, S, Madamwar, D, Singh, T.P. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the novel 14kDa fragment of alpha-subunit of phycoerythrin from the starving cyanobacterium Phormidium tenue.
J.Struct.Biol., 171, 2010
|
|
9AAT
 
 | |
9ANT
 
 | | ANTENNAPEDIA HOMEODOMAIN-DNA COMPLEX | | Descriptor: | ANTENNAPEDIA HOMEODOMAIN, DNA (5'-D(*AP*GP*AP*AP*AP*GP*CP*CP*AP*TP*TP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*CP*TP*AP*AP*TP*GP*GP*CP*TP*TP*TP*C)-3'), ... | | Authors: | Fraenkel, E, Pabo, C.O. | | Deposit date: | 1998-07-02 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparison of X-ray and NMR structures for the Antennapedia homeodomain-DNA complex.
Nat.Struct.Biol., 5, 1998
|
|
9ATC
 
 | | ATCASE Y165F MUTANT | | Descriptor: | ASPARTATE TRANSCARBAMOYLASE, ZINC ION | | Authors: | Ha, Y, Allewell, N.M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-07-26 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Intersubunit hydrogen bond acts as a global molecular switch in Escherichia coli aspartate transcarbamoylase.
Proteins, 33, 1998
|
|
8WWO
 
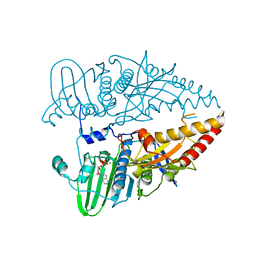 | |
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRR
 
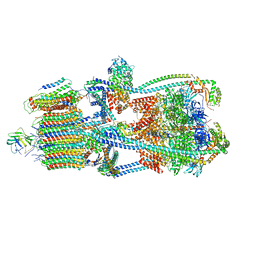 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRY
 
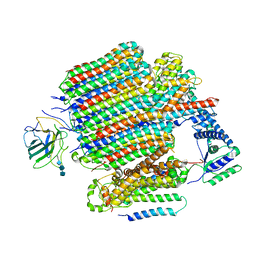 | | V0-only V-ATPase in synaptophysin gene knock-out mouse brain isolated synaptic vesicles | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-12 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-07 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 631, 2024
|
|
8WL9
 
 | |
8WL7
 
 | |
5EZT
 
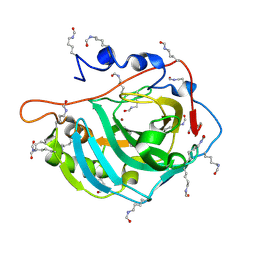 | | Peracetylated Bovine Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase 2, ZINC ION | | Authors: | Whitesides, G.M, Kang, K, Choi, J.-M, Fox, J.M. | | Deposit date: | 2015-11-26 | | Release date: | 2016-07-20 | | Last modified: | 2016-07-27 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Acetylation of Surface Lysine Groups of a Protein Alters the Organization and Composition of Its Crystal Contacts.
J.Phys.Chem.B, 120, 2016
|
|
