3ZO7
 
 | | Crystal structure of ClcFE27A with substrate | | Descriptor: | (2S)-2-chloranyl-2-[(2R)-5-oxidanylidene-2H-furan-2-yl]ethanoic acid, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-20 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.224 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
3WZD
 
 | | KDR in complex with ligand lenvatinib | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, ... | | Authors: | Okamoto, K, Ikemori_Kawada, M, Inoue, A, Matsui, J. | | Deposit date: | 2014-09-24 | | Release date: | 2015-05-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Distinct binding mode of multikinase inhibitor lenvatinib revealed by biochemical characterization.
ACS MED.CHEM.LETT., 6, 2015
|
|
2W6C
 
 | | ACHE IN COMPLEX WITH A BIS-(-)-NOR-MEPTAZINOL DERIVATIVE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3R)-3-ethyl-1-{9-[(3S)-3-ethyl-3-(3-hydroxyphenyl)azepan-1-yl]nonyl}azepan-3-yl]phenol, ... | | Authors: | Paz, A, Xie, Q, Greenblatt, H.M, Fu, W, Tang, Y, Silman, I, Qiu, Z, Sussman, J.L. | | Deposit date: | 2008-12-18 | | Release date: | 2009-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The Crystal Structure of a Complex of Acetylcholinesterase with a Bis-(-)-Nor-Meptazinol Derivative Reveals Disruption of the Catalytic Triad.
J.Med.Chem., 52, 2009
|
|
2VZG
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD2 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
2WFZ
 
 | | NON-AGED CONJUGATE OF TORPEDO CALIFORNICA ACETYLCHOLINESTERASE WITH SOMAN | | Descriptor: | (1R)-1,2,2-TRIMETHYLPROPYL (S)-METHYLPHOSPHINATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Sanson, B, Nachon, F, Colletier, J.P, Froment, M.T, Toker, L, Greenblatt, H.M, Sussman, J.L, Ashani, Y, Masson, P, Silman, I, Weik, M. | | Deposit date: | 2009-04-15 | | Release date: | 2009-08-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystallographic Snapshots of Nonaged and Aged Conjugates of Soman with Acetylcholinesterase, and of a Ternary Complex of the Aged Conjugate with Pralidoxime.
J.Med.Chem., 52, 2009
|
|
4D8Z
 
 | | Crystal structure of B. anthracis DHPS with compound 24 | | Descriptor: | (3R)-3-(7-amino-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
4D9Q
 
 | |
4D34
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)-N-(3- fluorophenethyl)ethan-1-amine | | Descriptor: | 2-(3-fluorophenyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}ethanamine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
2VT6
 
 | | Native Torpedo californica acetylcholinesterase collected with a cumulated dose of 9400000 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, CHLORIDE ION, ... | | Authors: | Colletier, J.P, Bourgeois, D, Sanson, B, Fournier, D, Sussman, J.L, Silman, I, Weik, M. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Shoot-and-Trap: Use of Specific X-Ray Damage to Study Structural Protein Dynamics by Temperature-Controlled Cryo-Crystallography.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4D8O
 
 | |
4D8H
 
 | | Crystal structure of Symfoil-4P/PV2: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 2 (6xLeu / PV1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo protein | | Authors: | Blaber, M, Longo, L. | | Deposit date: | 2012-01-10 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4D8L
 
 | | Crystal structure of the 2-pyrone-4,6-dicarboxylic acid hydrolase from sphingomonas paucimobilis | | Descriptor: | 2-pyrone-4,6-dicarbaxylate hydrolase | | Authors: | Malashkevich, V.N, Toro, R, Bonanno, J, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2012-01-10 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Catalytic Mechanism of LigI: Insight into the Amidohydrolase Enzymes of cog3618 and Lignin Degradation.
Biochemistry, 51, 2012
|
|
4D8V
 
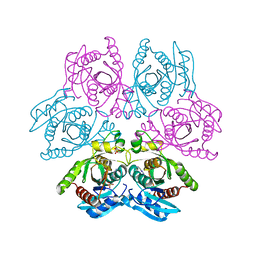 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis at pH 4.2 | | Descriptor: | ADENINE, Purine nucleoside phosphorylase deoD-type, SULFATE ION | | Authors: | Santos, C.R, Meza, A.N, Martins, N.H, Giuseppe, P.O, Murakami, M.T. | | Deposit date: | 2012-01-11 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
4D9R
 
 | |
2B2E
 
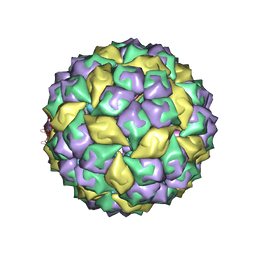 | | RNA stemloop from bacteriophage MS2 complexed with an N87S,E89K mutant MS2 capsid | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP*UP*AP*CP*CP*CP*AP*UP*GP*U)-3', Coat protein | | Authors: | Horn, W.T, Tars, K, Grahn, E, Helgstrand, C, Baron, A.J, Lago, H, Adams, C.J, Peabody, D.S, Phillips, S.E.V, Stonehouse, N.J, Liljas, L, Stockley, P.G. | | Deposit date: | 2005-09-19 | | Release date: | 2006-05-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of RNA binding discrimination between bacteriophages Qbeta and MS2
Structure, 14, 2006
|
|
2W0P
 
 | |
4D8D
 
 | |
4D9T
 
 | | Rsk2 C-terminal Kinase Domain with inhibitor (E)-methyl 3-(4-amino-7-(3-hydroxypropyl)-5-p-tolyl-7H-pyrrolo[2,3-d]pyrimidin-6-yl)-2-cyanoacrylate | | Descriptor: | Ribosomal protein S6 kinase alpha-3, SODIUM ION, methyl (2S)-3-{4-amino-7-[(1E)-3-hydroxyprop-1-en-1-yl]-5-(4-methylphenyl)-7H-pyrrolo[2,3-d]pyrimidin-6-yl}-2-cyanopropanoate | | Authors: | Serafimova, I.M, Pufall, M.A, Krishnan, S, Duda, K, Cohen, M.S, Maglathlin, R.L, McFarland, J.M, Miller, R.M, Frodin, M, Taunton, J. | | Deposit date: | 2012-01-12 | | Release date: | 2012-04-25 | | Last modified: | 2012-05-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversible targeting of noncatalytic cysteines with chemically tuned electrophiles.
Nat.Chem.Biol., 8, 2012
|
|
4D9Y
 
 | | The crystal structure of Chelerythrine bound to DNA d(CGTACG) | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, CALCIUM ION, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Ferraroni, M, Bazzicalupi, C, Gratteri, P, Bilia, A.R. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of Chelerythrine bound to DNA d(CGTACG)
To be Published
|
|
4D37
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-{[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methyl}-2-[2-(1H-imidazol-1-yl) pyrimidin-4-yl]ethanamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2015-03-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4D8K
 
 | |
4D9P
 
 | | Crystal structure of B. anthracis DHPS with compound 17 | | Descriptor: | (3R)-3-(7-amino-1-methyl-4,5-dioxo-1,4,5,6-tetrahydropyrimido[4,5-c]pyridazin-3-yl)butanoic acid, Dihydropteroate Synthase, SULFATE ION | | Authors: | Hammoudeh, D, Lee, R.E, White, S.W. | | Deposit date: | 2012-01-11 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structure-Based Design of Novel Pyrimido[4,5-c]pyridazine Derivatives as Dihydropteroate Synthase Inhibitors with Increased Affinity.
Chemmedchem, 7, 2012
|
|
4DA0
 
 | | Crystal structure of the hexameric purine nucleoside phosphorylase from Bacillus subtilis in complex with 2'-deoxyguanosine | | Descriptor: | 2'-DEOXY-GUANOSINE, CHLORIDE ION, Purine nucleoside phosphorylase deoD-type | | Authors: | Martins, N.H, Giuseppe, P.O, Meza, A.N, Murakami, M.T. | | Deposit date: | 2012-01-12 | | Release date: | 2012-09-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Insights into phosphate cooperativity and influence of substrate modifications on binding and catalysis of hexameric purine nucleoside phosphorylases.
Plos One, 7, 2012
|
|
2WFO
 
 | | Crystal structure of Machupo virus envelope glycoprotein GP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCOPROTEIN 1 | | Authors: | Bowden, T.A, Crispin, M, Graham, S.C, Harvey, D.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2009-04-09 | | Release date: | 2009-06-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Unusual Molecular Architecture of the Machupo Virus Attachment Glycoprotein.
J.Virol., 83, 2009
|
|
4D85
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BVI151 | | Descriptor: | (3R,4S,5S)-3-[(3-tert-butylbenzyl)amino]-5-[(4,4,7'-trifluoro-1',2'-dihydrospiro[cyclohexane-1,3'-indol]-5'-yl)methyl]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, IODIDE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
